7. Signal sequences initiate translocation
8.7 Signal sequences initiate translocation |
| Key terms defined in this section |
| Signal sequence is the region of a protein (usually N-terminal) responsible for co-translational insertion into membranes of the endoplasmic reticulum. |
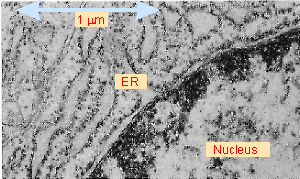 |
Figure 8.20 The endoplasmic reticulum consists of a highly folded sheet of membranes that extends from the nucleus. The small objects attached to the outer surface of the membranes are ribosomes. Photograph kindly provided by Lelio Orci. |
There is a common starting point for proteins that associate with, or pass through, the reticuloendothelial system of membranes. These proteins can associate with the membrane only while they are being synthesized. The ribosomes synthesizing these proteins become associated with the endoplasmic reticulum, enabling the nascent protein to be co-translationally transferred to the membrane. Regions in which ribosomes are associated with the ER are sometimes called the "rough ER," in contrast with the "smooth ER" regions that lack associated polysomes and which have a tubular rather than sheet-like appearance. Figure 8.20 shows ribosomes in the act of transferring nascent proteins to ER membranes.
The proteins synthesized at the rough endoplasmic reticulum pass from the ribosome directly to the membrane. From the endoplasmic reticulum membranes, the proteins are transferred to the Golgi apparatus, and then are directed to their ultimate destination, such as the lysosome or secretory vesicle or plasma membrane. (This is the subject of 25 Protein trafficking.)
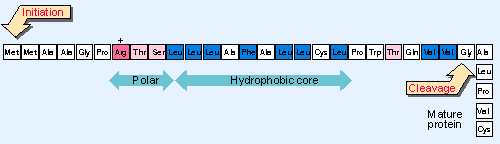 |
Figure 8.21 The signal sequence of bovine growth hormone consists of the N-terminal 29 amino acids and has a central highly hydrophobic region, preceded or flanked by regions containing polar amino acids. |
Co-translational insertion is directed by a signal sequence. Usually this is a cleavable leader sequence of the 15-30 N-terminal amino acids. At or close to the N-terminus are several polar residues, and within the leader is a hydrophobic core consisting exclusively or very largely of hydrophobic amino acids. There is no other conservation of sequence. Figure 8.21 gives an example.
Like the leader sequence of proteins targeted to organelles, the signal sequence is both necessary and sufficient to sponsor transfer of any attached polypeptide into the target membrane. A signal sequence added to the N-terminus of a globin protein, for example, causes it to be secreted through cellular membranes instead of remaining in the cytosol. However, the action of the signal sequence in co-translational transfer is different from that of the leader in post-translational transfer.
The signal sequence provides the connection that enables the ribosomes to attach to the membrane. There is no intrinsic difference between free ribosomes (synthesizing proteins in the cytosol) and ribosomes that are attached to the ER. A ribosome starts synthesis of a protein without knowing whether the protein will be synthesized in the cytosol or transferred to a membrane. It is the synthesis of a signal sequence that causes the ribosome to associate with a membrane.
Protein translocation can be divided into two general stages: first ribosomes carrying nascent polypeptides associate with the membranes; and then the nascent chain is transferred to the channel and translocates through it.
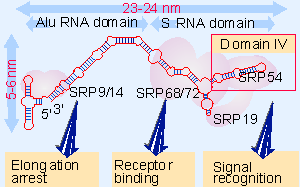 |
Figure 8.23 The two domains of the 7S RNA of the SRP are defined by its relationship to the Alu sequence. Five of the six proteins bind directly to the 7S RNA. Each function of the SRP is associated with a particular protein(s). |
The attechment of ribosomes to membranes requires the signal recognition particle (SRP). This is an 11S ribonucleoprotein complex, containing 6 proteins (total mass 240 kD) and a small (305 base, 100 kD) 7S RNA (see Figure 8.23). The 7S RNA provides the structural backbone of the particle; the individual proteins do not assemble in its absence (Blobel and Dobberstein, 1975; Walter and Blobel, 1981; Walter and Blobel, 1982).
The SRP has two important abilities:
- It can bind to the signal sequence of nascent secretory proteins.
- And it can bind to a protein (the SRP receptor) located in the membrane.
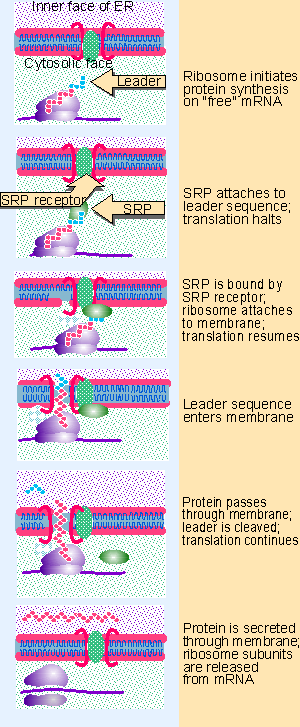 |
Figure 8.22 Ribosomes synthesizing secretory proteins are attached to the membrane via the signal sequence on the nascent polypeptide. Animated figure |
The SRP and SRP receptor function catalytically to transfer a ribosome carrying a nascent protein to the membrane. The first step is the recognition of the signal sequence by the SRP. Then the SRP binds to the SRP receptor and the ribosome binds to the membrane. The stages of translation of membrane proteins are summarized in Figure 8.22.
The role of the SRP receptor in protein translocation is transient. When the SRP binds to the signal sequence, it arrests translation. This usually happens when ~70 amino acids have been incorporated into the polypeptide chain (at this point the 25 residue leader has become exposed, with the next ~40 amino acids still buried in the ribosome).
Then when the SRP binds to the SRP receptor, the SRP releases the signal sequence. The ribosome becomes bound by another component of the membrane. At this point, translation can resume. When the ribosome has been passed on to the membrane, the role of SRP and SRP receptor has been played. They now recycle, and are free to sponsor the association of another nascent polypeptide with the membrane (for review see Walter and Johnson, 1994).
This process may be needed to control the conformation of the protein. If the nascent protein were released into the cytoplasm, it could take up a conformation in which it might be unable to traverse the membrane. The ability of the SRP to inhibit translation while the ribosome is being handed over to the membrane is therefore important in preventing the protein from being released into the aqueous environment.
The 7S RNA of the SRP particle is divided into two parts. The 100 bases at the 5′ end and 45 bases at the 3′ end are closely related to the sequence of Alu RNA, a common mammalian small RNA. They therefore define the Alu domain. The remaining part of the RNA comprises the S domain.
Different parts of the SRP structure depicted in Figure 8.23 have separate functions in protein targeting. SRP54 can be crosslinked to the signal sequence of a nascent protein; it is directly responsible for recognition of the substrate protein. SRP54 is a GTPase, and the cleavage of GTP is probably needed to insert the signal sequence into the channel. The SRP68-SRP72 dimer binds to the central region of the RNA; it is needed for recognizing the SRP receptor. The SRP9-SRP14 dimer binds at the other end of the molecule; it is responsible for elongation arrest (Siegel and Walter, 1988).
The SRP receptor is a dimer containing subunits SRα (72 kD) and SRβ (30 kD). The amino-terminal end of the large subunit is anchored in the endoplasmic reticulum. The bulk of the protein protrudes into the cytosol. A large part of the sequence of the cytoplasmic region of the protein resembles a nucleic acid-binding protein, with many positive residues. This suggests the possibility that the SRP receptor recognizes the 7S RNA in the SRP. Both subunits of the SRP receptor have GTP-binding motifs; GTP hydrolysis is required to release the SRP from its receptor.
There is a counterpart to SRP in bacteria, although it contains fewer components. E. coli contains a 4.5S RNA that associates with ribosomes and is homologous to the 7S RNA of the SRP. It associates with two proteins: Ffh is homologous to SRP54, and FtsY is homologous to the α subunit of the SRP receptor. In fact, FtsY replaces the functions of both the α and β SRP subunits; its N-terminal domain substitutes for SRPβ (which is an integral membrane protein) in membrane targeting, and the C-terminal domain interacts with the target protein.The role of this complex is more limited than that of SRP-SRP receptor. It is probably required to keep some (but not all) secreted proteins in a conformation that enables them to interact with the secretory apparatus. This could be the original connection between protein synthesis and secretion; in eukaryotes the SRP has acquired the additional roles of causing translational arrest and targeting to the membrane.
Why should the SRP have an RNA component? The answer must lie in the evolution of the SRP: it must have originated very early in evolution, in an RNA-dominated world, presumably in conjunction with a ribosome whose functions were mostly carried out by RNA The crystal structure of the complex between the protein-binding domain of 4.5S RNA and the RNA-binding domain of Ffh suggests that RNA continues to play a role in the function of SRP.
The 4.5S RNA has a region (domain IV) that is very similar to domain IV in 7S RNA (see Figure 8.23). Ffh consists of three domains (N, G, and M). The the M domain (named for a high content of methionines) performs the key binding functions (Keenan et al., 1998). It has a hydrophobic pocket that binds the signal sequence in a target protein. Next to the pocket is a helix-turn-helix motif that is typical of DNA-binding proteins (see 11.7 Repressor binds cooperatively at each operator using a helix-turn-helix motif).
Th crystal structure shows that the helix-loop-helix of the M domain binds to a duplex region of the 4.5S RNA in domain IV (Batey, et al., 2000). The negatively charged backbone of the RNA is adjacent to the hydrophobic pocket. This raises the possibility that a signal sequence actually binds to both the protein and RNA components of the SRP. The positively charged sequences that start the signal sequence (see Figure 8.21) could interact with the RNA, while the hydrophobic region of the signal sequence could sit in the pocket.
The signal peptide is cleaved from a translocating protein by a complex of 5 proteins called the signal peptidase. The complex is several times more abundant than the SRP and SRP receptor. Its amount is equivalent roughly to the amount of bound ribosomes, suggesting that it functions in a structural capacity. It is located on the lumenal face of the ER membrane, which implies that the entire signal sequence must cross the membrane before the cleavage event occurs.
This section updated 4-18-2000
| Reviews | |
| Walter, P. and Johnson, A. E. (1994). Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Ann. Rev. Cell Biol. 10, 87-119. | |
| Research | |
| Batey, R. T., Rambo, R. P., Lucast, L., Rha, B., and Doudna, J. A. (2000). Crystal structure of the ribonucleoprotein core of the signal recognition particle.. Science 287, 1232-1239. | |
| Blobel, G. and Dobberstein, B. (1975). Transfer of proteins across the membrane I Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67, 835-851. | |
| Keenan, R. J., Freymann, D. M., Walter, P., and Stroud, R. M. (1998). Crystal structure of the signal sequence-binding subunit of the signal recognition particle.. Cell 94, 181-191. | |
| Siegel, V. and Walter, P. (1988). Each of the activities of SRP is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell 52, 39-49. | |
| Walter, P. and Blobel, G. (1981). Translocation of proteins across the ER III SRP causes signal sequence and site specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 91, 557-561. | |
| Walter, P. and Blobel, G. (1982). Signal recognition particle contains a 7S RNA essential for protein translocation across the ER. Nature 299, 691-698. | |