8. The translocon forms a pore
8.8 The translocon forms a pore |
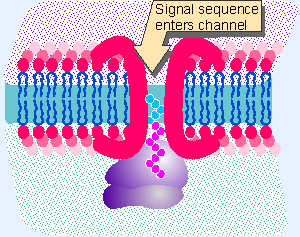 |
Figure 8.24 Does a signal sequence enter an aqueous tunnel created by resident ER membrane proteins? |
There is a basic problem in passing a (largely) hydrophilic protein through a hydrophobic membrane. The energetics of the interaction between the charged protein and the hydrophic lipids are highly unfavorable. However, a protein in the process of translocation across the ER membrane can be extracted by denaturants that are effective in an aqueous environment. The same denaturants do not extract proteins that are resident components of the membrane. This suggests the model for translocation illustrated in Figure 8.24, in which proteins of the ER membrane form an aqueous channel through the bilayer. A translocating protein moves through this channel, interacting with the resident proteins rather than with the lipid bilayer.
The channel through the membrane is called the translocon. Its components have been identified in two ways. Resident ER membrane proteins that are crosslinked to translocating proteins are potential subunits of the channel. And sec mutants in yeast (named because they fail to secrete proteins) include a class that cause precursors of secreted or membrane proteins to accumulate in the cytosol. These approaches together identified the Sec61 complex, which consists of three transmembrane proteins: Sec61α,β,γ. When this complex is incorporated into artificial membranes together with the SRP receptor, it can support translocation of some nascent proteins. Other nascent proteins require the presence of an additional component, TRAM, which is a major protein that becomes crosslinked to a translocating nascent chain. TRAM stimulates the translocation of all proteins.
Sec61 is the major component of the translocon. In detergent (which provides a hydrophobic milieu that mimics the effect of a surrounding membrane), Sec61 forms cylindrical oligomers with a diameter of ~85Å and a central pore of ~20Å. Each oligomer consists of 3-4 heterotrimers (Hanein et al., 1996).
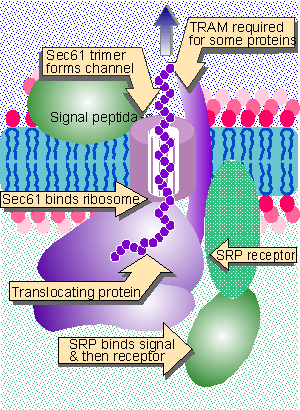 |
Figure 8.25 The translocon consists of SRP, SRP receptor, Sec61, TRAM, and signal peptidase. |
The components of the translocon and their functions are summarized in Figure 8.25. The simplicity of this system makes several important points. We visualize Sec61 as forming the channel and also as interacting with the ribosome. When the signal sequence enters the translocon, the ribosome attaches to Sec61, forming a seal so that the pore is not exposed to the cytosol. Cleavage of the signal peptide does not occur in this system, and therefore cannot be necessary for translocation per se. In this system, components on the lumenal side of the membrane are not needed for translocation. Of course, the efficiency of the in vitro system is relatively low. Additional components could be required in vivo to achieve efficient transfer or to prevent other cellular proteins from interfering with the process (Gorlich and Rapoport, 1993; for review see Walter and Lingappa, 1986; Rapoport et al., 1996).
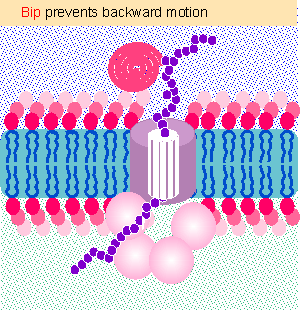 |
Figure 8.26 BiP acts as a ratchet to prevent backward diffusion of a translocating protein. Animated figure |
A more complex apparatus is required in certain cases in which a protein is inserted into a membrane post-translationally. The same Sec61 complex forms the channel, but four other Sec proteins are also required, and in addition the chaperone BiP (a member of the Hsp70 class) and a supply of ATP are required on the lumenal side of the membrane. Figure 8.26 shows that BiP behaves as ratchet (Matlack et al., 1999). In the absence of BiP, Brownian motion allows the protein to slip back into the cytosol. But BiP grabs the protein as it exits the pore into the endoplasmic reticulum. This stops the protein from moving backward. BiP does not pull the protein through; it just stops it from sliding back. (The reason why BiP is required for posttranslational translocation but not for cotranslational translocation may be that a newly synthesized protein is continuously extruded from the ribosome and therefore cannot slip backward.)
Is the channel a preexisting structure (as implied in the figure) or might it be assembled in response to the association of a hydrophobic signal sequence with the lipid bilayer? Channels can be detected by their ability to allow the passage of ions (measured as a localized change in electrical conductance). Ion-conducting channels can be detected in the ER membrane, and their state depends on protein translocation.
A channel opens when a nascent polypeptide is transferred from a ribosome to the ER membrane. The translocating protein fills the channel completely, so ions cannot pass through during translocation. But if the protein is released by treatment with puromycin, then the channel becomes freely permeable. If the ribosomes are removed from the membrane, the channel closes, suggesting that the open state requires the presence of the ribosome. This suggests that the channel is a permanent structure that provides an aqueous environment in which the protein may pass through the membrane. Measurements of the abilities of fluorescence quenching agents of different sizes to enter the channel suggest that it is large, with an internal diameter of 40-60Å. This is much larger than the diameter of an extended α-helical stretch of protein. It is also larger than the pore seen in direct views of the channel; this discrepancy remains to be explained (Simon and Blobel, 1991).
The aqueous environment of an amino acid in a protein can be measured by incorporating variant amino acids that have photoreactive residues. The fluorescence of these residues indicates whether they are in an aqueous or hydrophobic environment. Experiments with such probes show that when the signal sequence is first synthesized in the ribosome, it is in an aqueous state, but is not accessible to ions in the cytosol. It remains in the aqueous state throughout its interaction with a membrane. This suggests that the translocating protein travels directly from an enclosed tunnel in the ribosome into an aqueous channel in the membrane.
In fact, access to the pore is controlled (or "gated") on both sides of the membrane. Before attachment of the ribosome, the pore is closed on the lumenal side. When the ribosome attaches, it seals the pore on the cytosolic side. When the nascent protein reaches a length of ~70 amino acids, that is, probably when it extends fully across the channel, the pore opens on the lumenal side. So at all times, the pore is closed on one side or the other, maintaining the ionic integrities of the separate compartments (Crowley, 1994; Liao et al., 1997).
Several important activities occur within the endoplasmic reticulum. Proteins move through the ER en route to a variety of destinations (see 25 Protein trafficking). They are glycosylated and folded into their final conformations. The ER provides a "quality control" system in which misfolded proteins are identified and degraded. However, the degradation itself does not necessarily occur in the ER, but may require the protein to be exported back to the cytosol.
The first indication for cytosolic involvement in the degradation of ER proteins was provided by evidence for the involvement of the proteasome, a large protein aggregate with several proteolytic activities. Inhibitors of the proteasome prevent the degradation of aberrant ER proteins. Proteins are marked for cleavage by the proteasome when they are modified by the addition of ubiquitin, a small polypeptide chain. Ubiquitination and proteasomal activities are discussed later in this chapter; the important point to note now is that both are found in the cytosol (with a minor proportion in the nucleus).
Transport from the ER back into the cytosol occurs by a reversal of the usual process of import. The Sec61 translocon is used. The conditions are different; for example, the translocon is not associated with a ribosome. We do not know how the channel is opened to allow insertion of the protein on the ER side. Special components are presumably involved. In one particular case, human cytomegalovirus (CMV) codes for cytosolic proteins that destroy newly synthesized MHC class I (cellular major histocompatibility complex) proteins. This requires a viral protein product (US2), which is a membrane protein that functions in the ER. It interacts with the MHC proteins and probably conveys them into the translocon for reverse translocation.
The system involved in the degradation of aberrant ER proteins can be identified by mutations (in yeast) that lead to accumulation of aberrant proteins. Usually a protein that misfolds (produced by a mutated gene) is degraded instead of being transported through the ER. Yeast mutants that cannot degrade the substrate fall into two classes: some identify components of the proteolytic apparatus, such as the enzymes involved in ubiquitination; other identify components of the transport apparatus, including Sec61, BiP, and Sec63. There is also a protein in the ER membrane that functions on the cytosolic side to localize the ubiquitination enzymes at the translocon. In fact, retrograde transport into the cytosol cannot occur in the absence of this protein, which suggests that there is a mechanical link between retrograde transport and degradation (Wiertz et al., 1996).
| Reviews | |
| Rapoport, T. A., Jungnickel, B., and Kutay, U. (1996). Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Ann. Rev. Biochem 65, 271-303. | |
| Walter, P. and Lingappa, V. (1986). Mechanism of protein translocation across the endoplasmic reticulum membrane. Ann. Rev. Cell Biol. 2, 499-516. | |
| Research | |
| Crowley, K. S. (1994). Secretory proteins move through the ER membrane via an aqueous, gated pore. Cell 78, 461-471. | |
| Gorlich, D. and Rapoport, T. A. (1993). Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75, 615-630. | |
| Hanein, D. et al. (1996). Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell 87, 721-732. | |
| Liao, S. et al. (1997). Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell 90, 31-41. | |
| Matlack, K. E. S., Misselwitz, B., Plath, K., and Rapoport, T. A. (1999). BiP acts as a molecular ratchet during posttranslational transport of prepro-a factor across the ER membrane.. Cell 97, 553-564. | |
| Simon, S. M. and Blobel, G. (1991). A protein-conducting channel in the endoplasmic reticulum. Cell 65, 371-380. | |
| Wiertz, E. J. H. J. et al. (1996). Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432-438. | |