11. Enhancers contain bidirectional elements that assist initiation
20.11 Enhancers contain bidirectional elements that assist initiation |
| Key terms defined in this section |
| Enhancer element is a cis-acting sequence that increases the utilization of (some) eukaryotic promoters, and can function in either orientation and in any location (upstream or downstream) relative to the promoter. |
We have considered the promoter so far as an isolated region responsible for binding RNA polymerase. But eukaryotic promoters do not necessarily function alone. In at least some cases, the activity of a promoter is enormously increased by the presence of an enhancer, which consists of another group of elements, but located at a variable distance from those regarded as comprising part of the promoter itself (665; for review see 219).
The concept that the enhancer is distinct from the promoter reflects two characteristics. The position of the enhancer relative to the promoter need not be fixed, but can vary substantially. And it can function in either orientation. Manipulations of DNA show that an enhancer can stimulate any promoter placed in its vicinity.
For operational purposes, it is sometimes useful to define the promoter as a sequence or sequences of DNA that must be in a (relatively) fixed location with regard to the startpoint. By this definition, the TATA box and other upstream elements are included, but the enhancer is excluded. This is, however, a working definition rather than a rigid classification.
Elements analogous to enhancers, called upstream activator sequences (UAS), are found in yeast. They can function in either orientation, at variable distances upstream of the promoter, but cannot function when located downstream. They have a regulatory role: in several cases the UAS is bound by the regulatory protein(s) that activates the genes downstream.
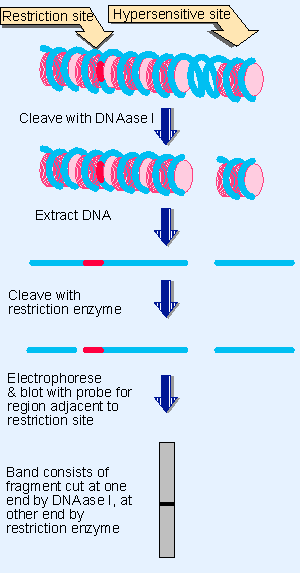 |
Figure 19.39 Indirect end-labeling identifies the distance of a DNAase hypersensitive site from a restriction cleavage site. The existence of a particular cutting site for DNAase I generates a discrete fragment, whose size indicates the distance of the DNAase I hypersensitive site from the restriction site. |
 |
Figure 19.40 The SV40 minichromosome has a nucleosome gap. Photograph kindly provided by Moshe Yaniv. |
An enhancer in the virus SV40 was one of the first to be characterized. It is located in a region of the genome that contains two identical sequences of 72 bp each, repeated in tandem ~200 bp upstream of the startpoint of a transcription unit. These 72 bp repeats lie in a region with an unusual chromatin structure, where the presence of a site hypersensitive to nuclease identifies a region in which DNA is more exposed than usual (see Figure 19.39 and Figure 19.40). Each 72 bp repeat contains a copy of the enhancer.
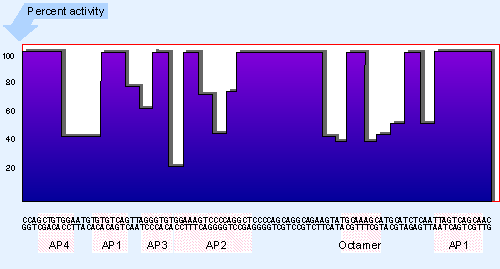 |
Figure 20.19 An enhancer contains several structural motifs. The histogram plots the effect of all mutations that reduce enhancer function to <75% of wild type. Binding sites for proteins are indicated below the histogram. |
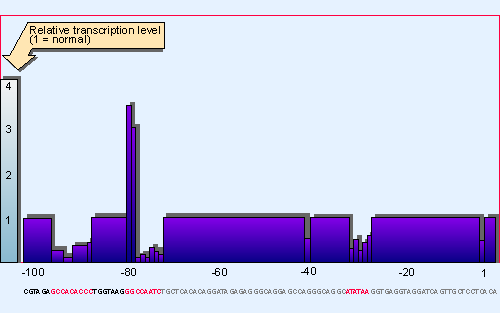 |
Figure 20.16 Saturation mutagenesis of the upstream region of the b-globin promoter identifies three short regions (centered at -30, -75, and -90) that are needed to initiate transcription. These correspond to the TATA, CAAT, |
A difference between the enhancer and a typical promoter is presented by the density of regulatory elements. Figure 20.19 summarizes the susceptibility of the SV40 enhancer to damage by mutation; and we see that a much greater proportion of its sites directly influences its function than is the case with the promoter analyzed in the same way in Figure 20.16. There is a corresponding increase in the density of protein-binding sites. Many of these sites are common elements in promoters; for example, AP1 and the octamer.
Enhancers often show redundancy in function. The SV40 enhancer can be separated into two halves; they function poorly as enhancers by themselves, but constitute an effective enhancer together or even when they are separated by introducing sequences between them. Mutations that inactivate the left element can be compensated by duplicating other regions in the enhancer. Although these regions are different in sequence, they appear to play similar roles, since an active enhancer can be created by the combination of a sufficient number of wild-type domains, irrespective of their types. Such redundancy is common in enhancers; the result is that multiple mutations are required, to eliminate more than one element, before an enhancer is inactivated. In the SV40 enhancer, no individual mutation decreases activity by as much as 10 .
Cellular enhancers have similar properties. An enhancer works upon the promoter that is nearest to it, but the enhancer may be either upstream or downstream of the promoter. Responsibility for tissue-specific transcription may lie with either a promoter or enhancer. A promoter may be specifically regulated, and a nearby enhancer used to increase the efficiency of initiation; or a promoter may lack specific regulation, but become active only when a nearby enhancer is specifically activated. An example is provided by immunoglobulin genes, which carry enhancers within the transcription unit. The immunoglobulin enhancers appear to be active only in the B lymphocytes in which the immunoglobulin genes are expressed. Such enhancers provide part of the regulatory network by which gene expression is controlled.
Reconstruction experiments in which the enhancer sequence is removed from the DNA and then is inserted elsewhere show that normal transcription can be sustained so long as it is present anywhere on the DNA molecule. If a β-globin gene is placed on a DNA molecule that contains an enhancer, its transcription is increased in vivo more than 200-fold, even when the enhancer is several kb upstream or downstream of the startpoint, in either orientation. We have yet to discover at what distance the enhancer fails to work.
How can an enhancer stimulate initiation at a promoter that can be located at apparently any distance away on either side of it? When enhancers were first discovered, several possibilities were considered for their action as elements distinctly different from promoters:
- An enhancer could change the overall structure of the template Xfor example, by influencing the density of supercoiling.
- It could be responsible for locating the template at a particular place within the cell Xfor example, attaching it to the nuclear matrix.
- An enhancer could provide an "entry site," a point at which RNA polymerase (or some other essential protein) associates with chromatin.
Now we take the view that enhancer function involves the same sort of interaction with the basal apparatus as the interactions sponsored by upstream promoter elements. Enhancers are modular, like promoters. Some elements are found in both enhancers and promoters. Some individual elements found in promoters share with enhancers the ability to function at variable distance and in either orientation. So the distinction between enhancers and promoters is blurred: enhancers might be viewed as containing promoter elements that are grouped closely together, with the ability to function at increased distances from the startpoint (666).
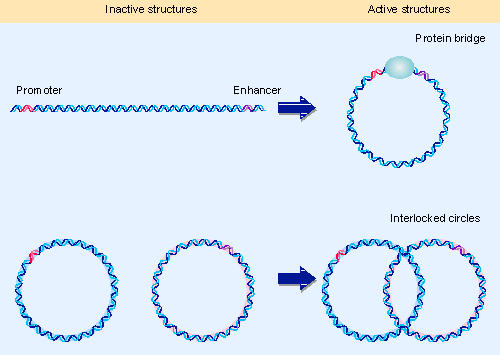 |
Figure 20.20 Figure 20.20 An enhancer may function by bringing proteins into the vicinity of the promoter. An enhancer does not act on a promoter at the opposite end of a long linear DNA, but becomes effective when the DNA is joined into a circle by a protein bridge. An enhancer and promoter on separate circular DNAs do not interact, but can interact when the two molecules are catenated. |
If the enhancer represents an extreme case of the ability to mix and match promoter elements, it might be considered a part of the promoter in a more distant location. The essential role of the enhancer may be to increase the concentration of transcription factors in the vicinity of the promoter (vicinity in this sense being a relative term). Two types of experiment illustrated in Figure 20.20 suggest that this is the case.
A fragment of DNA that contains an enhancer at one end and a promoter at the other is not effectively transcribed, but the enhancer can stimulate transcription from the promoter when they are connected by a protein bridge. Since structural effects, such as changes in supercoiling, could not be transmitted across such a bridge, this suggests that the critical feature is bringing the enhancer and promoter into close proximity (667).
A bacterial enhancer provides a binding site for the regulator NtrC, which acts upon RNA polymerase using promoters recognized by σ54. When the enhancer is placed upon a circle of DNA that is catenated (interlocked) with a circle that contains the promoter, initiation is almost as effective as when the enhancer and promoter are on the same circular molecule. But there is no initiation when the enhancer and promoter are on separated circles. Again this suggests that the critical feature is localization of the protein bound at the enhancer, to increase its chance of contacting a protein bound at the promoter.
If proteins bound at an enhancer several kb distant from a promoter interact directly with proteins bound in the vicinity of the startpoint, the organization of DNA must be flexible enough to allow the enhancer and promoter to be closely located. This requires the intervening DNA to be extruded as a large "loop." Such loops have been directly observed in the case of the bacterial enhancer.
The generality of enhancement is not yet clear. We do not know what proportion of cellular promoters require an enhancer to achieve their usual level of expression. Nor do we know how often an enhancer provides a target for regulation. Some enhancers are activated only in the tissues in which their genes function, but others could be active in all cells.
A difference between enhancers and promoters may be that an enhancer shows greater cooperativity between the binding of factors. A complex that assembles at the enhancer that responds to IFN (interferon) γassembles cooperatively to form a functional structure called the enhanceosome. Binding of the nonhistone protein HMGI(Y) bends the DNA into a structure that then binds several transcription factors (NF-κB, IRF, ATF-Jun). In contrast with the "mix and match" construction of promoters, all of these components are required to create an active structure at the enhancer. These components do not themselves directly bind to RNA polymerase, but they create a surface that binds a coactivating complex. The coactivator binds RNA polymerase II and recruits it to the pre-initiation complex of basal transcription factors that is assembling at the promoter. We discuss the function of coactivators in more detail later.
| Reviews | |
| 219: | Muller, M. M., Gerster, T., and Schaffner, W. (1988). Enhancer sequences and the regulation of gene transcription. Eur. J. Biochem. 176, 485-495. |
| Research | |
| 665: | Banerji, J., Rusconi, S., and Schaffner, W. (1981). Expression of b-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299-308. |
| 666: | Zenke, M. et al. (1986). Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 5, 387-397. |
| 667: | Mueller-Storm, H. P., Sogo, J. M., and Schaffner, W. (1989). An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell 58, 767-777. |
- Chapter IV How Consumers Think About Interactive Aspects of Web Advertising
- Chapter XII Web Design and E-Commerce
- Chapter XIII Shopping Agent Web Sites: A Comparative Shopping Environment
- Chapter XVI Turning Web Surfers into Loyal Customers: Cognitive Lock-In Through Interface Design and Web Site Usability
- Chapter XVII Internet Markets and E-Loyalty