12. Independent domains bind DNA and activate transcription
20.12 Independent domains bind DNA and activate transcription |
Transcription factors and other regulatory proteins require two types of ability:
- They recognize specific target sequences located in enhancers, promoters, or other regulatory elements that affect a particular target gene.
- Having bound to DNA, a transcription factor, or a positive regulatory protein, exercises its function by binding to other components of the transcription apparatus.
Can we characterize domains in the transcription factors that are responsible for these activities? Often a factor has separate domains that bind DNA and activate transcription. Each domain behaves as a separate module that functions independently when it is linked to a domain of the other type. The geometry of the overall transcription complex must allow the activating domain to contact the basal apparatus irrespective of the exact location and orientation of the DNA-binding domain.
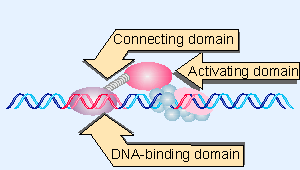 |
Figure 20.21 DNA-binding and activating functions in a transcription factor may comprise independent domains of the protein. |
Upstream promoter elements may be an appreciable distance from the startpoint, and in many cases may be oriented in either direction. Enhancers may be even farther away and always show orientation independence. This organization has implications for both the DNA and proteins. The DNA may be looped or condensed in some way to allow the formation of the transcription complex. And the domains of the transcription factor may be connected in a flexible way, as illustrated diagrammatically in Figure 20.21.
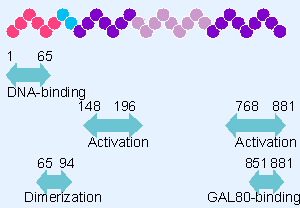 |
Figure 20.22 The GAL4 protein has independent regions that bind DNA, activate transcription (2 regions), dimerize, and bind the regulator GAL80. |
The modular nature of a transcriptional activator has been most thoroughly characterized for the example of the yeast activator GAL4, which controls the transcription of genes whose products are responsible for metabolizing galactose. Regulation is exercised at UASG (an upstream activating sequence). The GAL4 protein has three functions: it binds to a DNA sequence in the UASG; it activates transcription; and it binds another regulator protein, called GAL80. (GAL80 prevents GAL4 from activating transcription: GAL80 is released when galactose is present, thus allowing GAL4 to activate its target genes.) Figure 20.22 shows that each of these functions can be localized in a particular region of the GAL4 protein. In fact, GAL4 has two activating domains, each able to activate transcription. Another domain confers the ability to dimerize.
Binding to DNA naturally is prerequisite for activating transcription. But does activation depend on the particular DNA-binding domain?
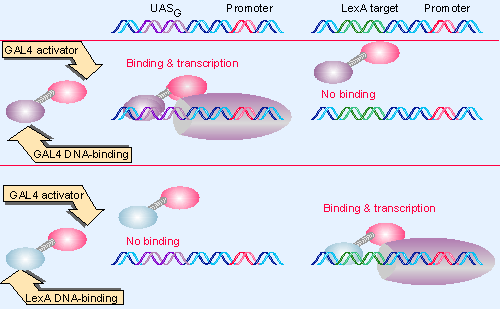 |
Figure 20.23 The ability of GAL4 to activate transcription is independent of its specificity for binding DNA. When the GAL4 DNA-binding domain is replaced by the LexA DNA-binding domain, the hybrid protein can activate transcription when a LexA operator is placed near a promoter. |
Figure 20.23 illustrates an experiment to answer this question. The bacterial repressor LexA has an N-terminal DNA-binding domain that recognizes a specific operator; binding of LexA at this operator represses the adjacent promoter. In a "swap" experiment, this sequence can be substituted for the DNA-binding domain of GAL4. The hybrid gene can then be introduced into yeast together with a target gene that contains either the UAS or a LexA operator.
An authentic GAL4 protein can activate a target gene only if it has a UAS. The LexA repressor by itself of course lacks the ability to activate either sort of target. The LexA-GAL4 hybrid can no longer activate a gene with a UAS, but it can now activate a gene that has a LexA operator!
This result fits the modular view of transcription activators. The DNA-binding domain serves to bring the protein into the right location. Precisely how or where it is bound to DNA is irrelevant, but, once it is there, the transcription-activating domain can play its role. According to this view, it does not matter whether the transcription-activating domain is brought to the vicinity of the promoter by recognition of a UAS via the DNA-binding domain of GAL4 or by recognition of a LexA operator via the LexA specificity module. The ability of the two types of module to function in hybrid proteins suggests that each domain of the protein folds independently into an active structure that is not influenced by the rest of the protein (for review see 230, 232).
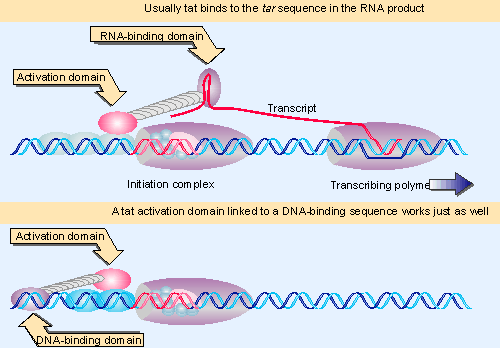 |
Figure 20.24 The activating domain of the tat protein of HIV can stimulate initiation if it is tethered in the vicinity by binding to the RNA product of a previous round of transcription. Activation is independent of the means |
The idea that transcription factors have independent domains that bind DNA and that activate transcription is reinforced by the ability of the tat protein of HIV to stimulate initiation without binding DNA at all. The tat protein binds to a region of secondary structure in the RNA product; the part of the RNA required for tat action is called the tar sequence. A model for the role of the tat-tar interaction in stimulating transcription is shown in Figure 20.24.
The tar sequence is located just downstream of the startpoint, so that when tat binds to tar, it is brought into the vicinity of the initiation complex. This is sufficient to ensure that its activation domain is in close enough proximity to the initiation complex. The activation domain interacts with one or more of the transcription factors bound at the complex in the same way as an upstream transcription factor. (Of course, the first transcript must be made in the absence of tat in order to provide the binding site.)
An extreme demonstration of the independence of the localizing and activating domains is indicated by some constructs in which tat was engineered so that the activating domain was connected to a DNA-binding domain instead of to the usual tar-binding sequence. When an appropriate target site is placed into the promoter, the tat activating-domain could activate transcription. This suggests that we should think of the DNA-binding (or in this case the RNA-binding) domain as providing a "tethering" function, whose main purpose is to ensure that the activating domain is in the vicinity of the initiation complex.
The notion of tethering is a more specific example of the general idea that initiation requires a high concentration of transcription factors in the vicinity of the promoter. This may be achieved when factors bind to enhancers in the general vicinity, when factors bind to upstream promoter components, or in an extreme case by tethering to the RNA product. The common requirement of all these situations is flexibility in the exact three dimensional arrangement of DNA and proteins. The principle of independent domains is common in transcriptional activators.
We might view the function of the DNA-binding domain as bringing the activating domain into the vicinity of the startpoint. This explains why the exact locations of DNA-binding sites can vary within the promoter.
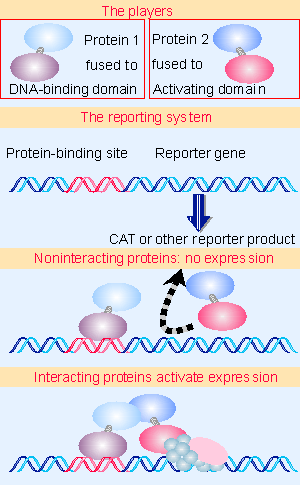 |
Figure 20.25 The two hybrid technique tests the ability of two proteins to interact by incorporating them into hybrid proteins where one has a DNA-binding domain and the other has a transcription-activating domain. Animated figure |
This model is the basis for an extremely useful assay for detecting protein interactions. In effect, we replace the connecting domain in Figure 20.21 with a protein-protein interaction. The principle is illustrated in Figure 20.25. We fuse one of the proteins to be tested to a DNA-binding domain. We fuse the other protein to a transcription-activating domain. (This is done by linking the appropriate coding sequences in each case and making synthetic proteins by expressing each hybrid gene.) If the two proteins that are being tested can interact with one another, the two hybrid proteins will interact. This is reflected in the name of the technique: the two hybrid assay (952). The protein with the DNA-domain binds to a reporter gene that has a simple promoter containing its target site. But it cannot activate the gene by itself. Activation occurs only if the second hybrid binds to the first hybrid to bring the activation domain to the promoter. Any reporter gene can be used where the product is readily assayed, and this technique has given rise to several automated procedures for rapidly testing protein-protein interactions.
The effectiveness of the technique dramatically illustrates the modular nature of proteins. Even when fused to another protein, the DNA-binding domain can bind to DNA and the transcription-activating domain can activate transcription. Correspondingly, the interaction ability of the two proteins being tested is not inhibited by the attachment of the DNA-binding or transcriptoion-activating domains. (Of course, there are some exceptions where these simple rules do not apply and interference between the domains of the hybrid protein prevents the technique from working.)
This section updated 4-12-2000
| Reviews | |
| 230: | Guarente, L. (1987). Regulatory proteins in yeast. Ann. Rev. Genet. 21, 425-452. |
| 232: | Ptashne, M. (1988). How eukaryotic transcriptional activators work. Nature 335, 683-689. |
| Research | |
| 952: | Fields, S. and Song, O. (1989). A novel genetic system to detect protein-protein interactions.. Nature 340, 245-246. |
- Enterprise Application Integration: New Solutions for a Solved Problem or a Challenging Research Field?
- Distributed Data Warehouse for Geo-spatial Services
- Data Mining for Business Process Reengineering
- Intrinsic and Contextual Data Quality: The Effect of Media and Personal Involvement
- Healthcare Information: From Administrative to Practice Databases