2. Condensing viral genomes into their coats
18.2 Condensing viral genomes into their coats |
| Key terms defined in this section |
| Capsid is the external protein coat of a virus particle. |
From the perspective of packaging the individual sequence, there is an important difference between a cellular genome and a virus. The cellular genome is essentially indefinite in size; the number and location of individual sequences can be changed by duplication, deletion, and rearrangement. So it requires a generalized method for packaging its DNA, insensitive to the total content or distribution of sequences. By contrast, two restrictions define the needs of a virus. The amount of nucleic acid to be packaged is predetermined by the size of the genome. And it must all fit within a coat assembled from a protein or proteins coded by the viral genes.
A virus particle is deceptively simple in its superficial appearance. The nucleic acid genome is contained within a capsid, a symmetrical or quasi-symmetrical structure assembled from one or only a few proteins. Attached to the capsid, or incorporated into it, are other structures, assembled from distinct proteins, and necessary for infection of the host cell.
The virus particle is tightly constructed. The internal volume of the capsid is rarely much greater than the volume of the nucleic acid it must hold. The difference is usually less than twofold, and often the internal volume is barely larger than the nucleic acid.
In its most extreme form, the restriction that the capsid must be assembled from proteins coded by the virus means that the entire shell is constructed from a single type of subunit. The rules for assembly of identical subunits into closed structures restrict the capsid to one of two types. The protein subunits stack sequentially in a helical array to form a filamentous or rod-like shape. Or they form a pseudospherical shell, a type of structure that conforms to a polyhedron with icosahedral symmetry. Some viral capsids are assembled from more than a single type of protein subunit, but although this extends the exact types of structures that can be formed, viral capsids still all conform to the general classes of quasi-crystalline filaments or icosahedrons (Caspar and Klug, 1962).
There are two types of solution to the problem of how to construct a capsid that contains nucleic acid:
- The protein shell can be assembled around the nucleic acid, condensing the DNA or RNA by protein-nucleic acid interactions during the process of assembly.
- Or the capsid can be constructed from its component(s) in the form of an empty shell, into which the nucleic acid must be inserted, being condensed as it enters.
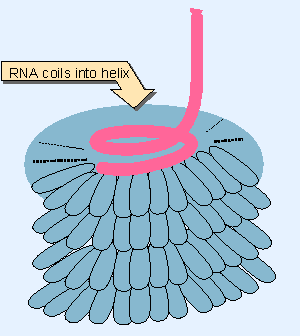 |
Figure 18.2 A helical path for TMV RNA is created by the stacking of protein subunits in the virion. |
The capsid is assembled around the genome for single-stranded RNA viruses. The principle of assembly is that the position of the RNA within the capsid is determined directly by its binding to the proteins of the shell. The best characterized example is TMV (tobacco mosaic virus). Assembly starts at a duplex hairpin that lies within the RNA sequence. From this nucleation center, it proceeds bidirectionally along the RNA, until reaching the ends. The unit of the capsid is a two-layer disk, each layer containing 17 identical protein subunits. The disk is a circular structure, which forms a helix as it interacts with the RNA. The RNA becomes coiled in a helical array on the inside of the protein shell, as illustrated in Figure 18.2 (Fraenkel-Conrat and Williams, 1955).
The spherical capsids of DNA viruses are assembled in a different way, as best characterized for the phages lambda and T4. In each case, an empty headshell is assembled from a small set of proteins. Then the duplex genome is inserted into the head, a process accompanied by a structural change in the capsid.
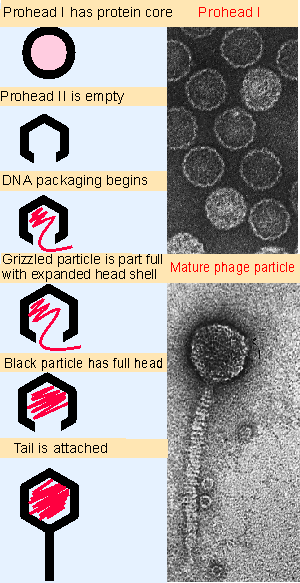 |
Figure 18.3 Maturation of phage lambda passes through several stages. The empty head changes shape and expands when it becomes filled with DNA. The electron micrographs show the particles at the start and end of the maturation pathway. Photographs kindly provided by A. F. Howatson. |
Figure 18.3 summarizes the assembly of lambda. It starts with a small headshell that contains a protein "core." This is converted to an empty headshell of more distinct shape. Then DNA packaging begins, the headshell expands in size though remaining the same shape, and finally the full head is sealed by addition of the tail.
Now a double-stranded DNA considered over short distances is a fairly rigid rod. Yet it must be compressed into a compact structure to fit within the capsid. We should like to know whether packaging involves a smooth coiling of the DNA into the head or requires abrupt bends.
Inserting DNA into a phage head involves two types of reaction: translocation and condensation. Both are energetically unfavorable.
Translocation is an active process in which the DNA is driven into the head by an ATP-dependent mechanism. One possibility is that the terminase enzymes that generate the ends of DNA (from longer precursor DNAs) could be involved in pushing it into the head. Another possibility is that the capsid protein(s) could pull the DNA in.
Little is known about the mechanism of condensation, except that the capsid contains "internal proteins" as well as DNA. One possibility is that they provide some sort of "scaffolding" onto which the DNA condenses. (This would be a counterpart to the use of the proteins of the shell in the plant RNA viruses.)
How specific is the packaging? It cannot depend on particular sequences, because deletions, insertions, and substitutions all fail to interfere with the assembly process. The relationship between DNA and the headshell has been investigated directly by determining which regions of the DNA can be chemically crosslinked to the proteins of the capsid. The surprising answer is that all regions of the DNA are more or less equally susceptible. This probably means that when DNA is inserted into the head, it follows a general rule for condensing, but the pattern is not determined by particular sequences (for review see Black, 1989).
These varying mechanisms of virus assembly all accomplish the same end: packaging a single DNA or RNA molecule into the capsid. However, some viruses have genomes that consist of multiple nucleic acid molecules. Reovirus contains ten double-stranded RNA segments, all of which must be packaged into the capsid. Specific sorting sequences in the segments may be required to ensure that the assembly process selects one copy of each different molecule in order to collect a complete set of genetic information.
Some plant viruses are multipartite: their genomes consist of segments, each of which is packaged into a different capsid. An example is alfalfa mosaic virus, which has four different single-stranded RNAs, each packaged independently into a coat comprising the same protein subunit. A successful infection depends on the entry of one of each type into the cell.
The four components of the virus exist as particles of different sizes. This means that the same capsid protein can package each RNA into its own characteristic particle. This is a departure from the packaging of a unique length of nucleic acid into a capsid of fixed shape. The assembly pathway of viruses whose capsids have only one authentic form may be diverted by mutations that cause the formation of aberrant monster particles in which the head is longer than usual. These mutations show that a capsid protein(s) has an intrinsic ability to assemble into a particular type of structure, but the exact size and shape may vary. Some of the mutations occur in genes that code for assembly proteins, which are needed for head formation, but are not themselves part of the headshell. Such ancillary proteins limit the options of the capsid protein so that it assembles only along the desired pathway. Comparable proteins are employed in the assembly of cellular chromatin (see 19 Nucleosomes).
| Reviews | |
| Black, L. W. (1989). DNA packaging in dsDNA bacteriophages. Ann. Rev. Immunol. 43, 267-292. | |
| Research | |
| Caspar, D. L. D. and Klug, A. (1962). Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27, 1-24. | |
| Fraenkel-Conrat, H. and Williams, R. C. (1955). Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Nat. Acad. Sci. USA 41, 690-698. | |
- The ROI of Lean Six Sigma for Services
- Seeing Services Through Your Customers Eyes-Becoming a customer-centered organization
- Success Story #3 Fort Wayne, Indiana From 0 to 60 in nothing flat
- Success Story #4 Stanford Hospital and Clinics At the forefront of the quality revolution
- First Wave Service Projects