11. The winglesswnt signaling pathway
29.10 Cell fate is determined by compartments that form by the blastoderm stage |
| Key terms defined in this section |
| Gap in DNA is the absence of one or more nucleotides in one strand of the duplex. |
A "fate map" of the embryo can be drawn at the blastoderm stage to identify regions in terms of the adult segments that will develop from the descendants of the embryonic cells. This is possible because, by this stage, cells have begun to acquire information about the pathways they will follow and the structures they will therefore form. This information derives from the maternal regulators described above, and then it is further refined by the actions of zygotic genes. The concept that is intrinsic in the fate map is that a region identified at blastoderm consists of a "compartment" of cells that will give rise specifically to a particular adult structure.
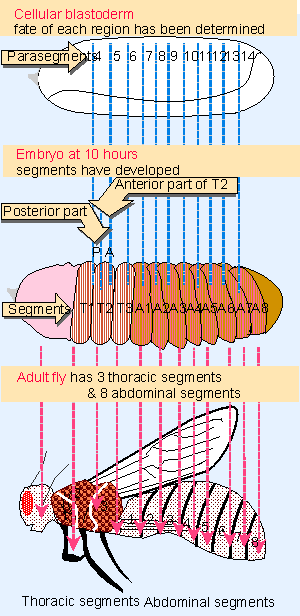 |
Figure 29.21 Drosophila development proceeds through formation of compartments that define parasegments and segments. |
We can consider the development of D. melanogaster in terms of the two types of unit depicted in Figure 29.21: the segment and parasegment.
- The segment is a visible morphological structure. The adult fly consists of a series of clearly demarcated segments, and the larva has a series of corresponding segments separated by grooves. We are concerned primarily with the three thoracic (T) and eight abdominal (A) segments, about whose development most is known. The pattern of segmental units is determined by blastoderm, when the main mass of the embryo is divided into a series of alternating anterior (A) and posterior (P) compartments. So a segment consists of an A compartment succeeded by a P compartment; segment A3, for example, consists of compartments A3A and A3P.
- Another type of classification originates earlier, when divisions can first be seen at gastrulation. The embryo can be divided into parasegments, each consisting of a P compartment succeeded by an A compartment. Parasegment 8, for example, consists of compartments A2P and A3A. In the 5 V6 hour embryo, shallow grooves on the surface separate the adjacent parasegments. When segments form at around 9 hours, the grooves deepen and move, so that each segmental boundary represents the center of a parasegment. So the anterior part of the segment is derived from one parasegment, and the posterior part of the segment is derived from the next parasegment. In effect, the segmental units are initially evident as P-A pairs in parasegments, and then are recognized as A-P pairs in segments.
How are these compartments defined during embryogenesis? The general nature of segmentation mutants suggests that the functions of segmentation genes are to establish "rules" by which segments form; a mutation changes a rule in such a way as to cause many or all segments to form improperly. The drastic consequences of segment malformation make these mutants embryonic lethals Xthey die at various stages before metamorphosis into adults (Nusslein-Vollhard and Wieschaus, 1980).
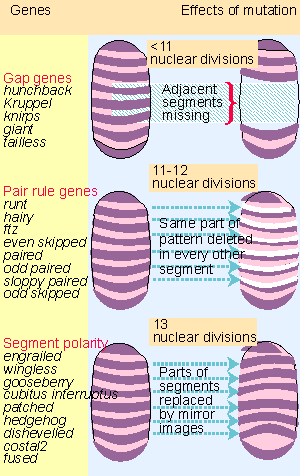 |
Figure 29.22 Segmentation genes affect the number of segments and fall into three groups. |
Probably ~30 loci are involved in segment formation. Figure 29.22 shows that they can be classified according to the size of the unit that they affect:
- Gap mutants have a group of several adjacent segments deleted from the final pattern. Four gap genes are involved in formation of the major body segments, and others are concerned with the head and tail structures.
- Pair-rule mutants have corresponding parts of the pattern deleted in every other segment. The afflicted segments may be even-numbered or odd-numbered. There are 8 pair-rule genes.
- Segment polarity mutants most often lose part of the P compartment of each segment, and it is replaced by a mirror image duplication of the A compartment. Some mutants cause loss of A compartments or middle segments. There are ~16 segment polarity genes.
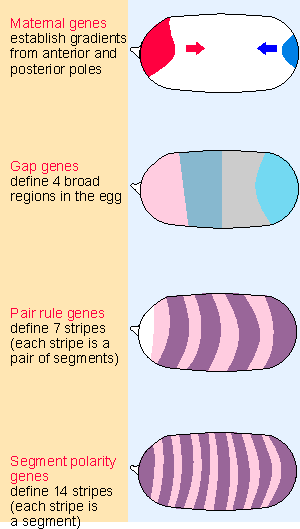 |
Figure 29.23 Maternal and segmentation genes act progressively on smaller regions of the embryo. |
These groups of genes are expressed at successive periods during development; and they define increasingly restricted regions of the egg, as can be seen from Figure 29.23. The maternal genes establish gradients from the anterior and posterior ends. The maternal gradients either activate or repress the gap genes, which are amongst the earliest to be transcribed following fertilization (following the 11th nuclear division); they divide the embryo into 4 broad regions. The gap genes regulate the pair-rule genes, which are transcribed slightly later; their target regions are restricted to pairs of segments. The pair-rule genes in turn regulate the segment polarity genes, which are expressed during the 13th nuclear division, and by now the target size is the individual segment.
Many of the maternal genes, the gap genes, and the pair-rule genes are regulators of transcription. Their effects may be either to activate or to repress transcription; in some cases, a given protein may activate some target genes and repress other target genes, depending on its level or the context. The genes in any one class regulate one another as well as regulating the genes of the next class. It is only when we reach the level of segment polarity genes that products are found that act in other ways; for example, secretion of a protein from one cell to influence its neighbor (for review see Ingham and Martinez-Arias, 1992; Scott and Carroll, 1987).
The principle that emerges from this analysis is that at each stage a small number of maternal, gap, and pair-rule regulator proteins is used in combinatorial associations to specify the pattern of gene expression in a particular region of the embryo.
The gap genes are controlled in two ways: they may respond directly to the bicoid morphogen; and they regulate one another. The 4 bands shown in Figure 29.23 are created in the following way. The most anterior band consists of hunchback protein, transcribed from the hunchback RNA whose synthesis was activated by bicoid. The next band consists of Kruppel protein; transcription of the Kruppel gene is activated by hunchback protein. The next two bands consist of knirps and giant proteins. Transcription of these genes is repressed by hunchback. They are expressed in the posterior part of the embryo because nanos has prevented the expression of hunchback there.
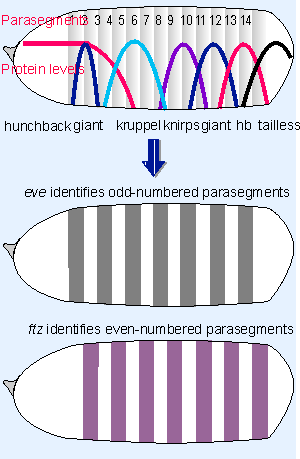 |
Figure 29.24 Expression of the gap genes defines adjacent regions of the embryo. The gap genes control the pair-rule genes, each of which is expressed in 7 stripes. |
Figure 29.24 examines the transition from the 4 band to the 7 striped stage in more detail. The detailed interactions among the gap proteins are determined by examining the pattern of the distribution of other gap proteins in a mutant lacking one particular gap protein. Hunchback plays an especially important role. It is expressed in a broad anterior region, with a gradient of decline in the middle of the embryo. High levels of hunchback repress Kruppel; this determines the anterior boundary of Kruppel expression, which rises just as hunchback falls off, in parasegment 3. But some level of hunchback is needed for Kruppel expression, so when the level of hunchback decreases further, Kruppel is turned off, around parasegment 5. In the same way, expression of giant responds to successive changes in the level of hunchback; and knirps expression requires the absence of hunchback.
The control is refined further by interactions among the proteins. The general principle is that one interaction may be required to express a protein in a particular region, and other interactions may be required to repress its expression at the boundaries. The effects are worked out by examining pairwise interactions. For example, over-expression of giant causes the Kruppel band to become much narrower, suggesting that giant contributes to repressing the boundaries of Kruppel. The posterior margins of knirps and giant are determined by the operation of the terminal system. Altogether, these interactions mean that, as we proceed along the egg from anterior to posterior, any particular position can be defined by the levels of the various gap proteins.
All the gap proteins are regulators of transcription, and in addition to regulating one another, they regulate the expression of the pair-rule genes that function at the next stage. Each pair-rule protein is found in a pattern of 7 "stripes" along the embryo, and Figure 29.24 shows the approximate positions that these stripes will take as the result of expression of the gap genes. (Of course, the parasegments have not developed yet, and are shown just to relate their positions to the protein distribution.) The 7 stripes of a pair-rule gene identify either all the odd-numbered parasegments (like eve) or all the even-numbered parasegments (like ftz). Two of the pair-rule genes, hairy and eve, are called primary pair-rule genes, because they are expressed first, and their pattern of expression influences the expression of the other pair-rule genes.
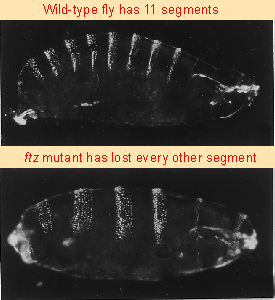 |
Figure 29.25 ftz mutants have half the number of segments present in wild-type. Photographs kindly provided by Walter Gehring. |
Recall that mutations in pair-rule genes delete half the segments. Figure 29.25 compares the segmentation patterns of wild-type and fushi tarazu (ftz) larvae. The mutant has only half the number of segments, because every other segment is missing.
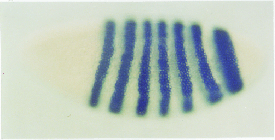 |
Figure 29.26 Transcripts of the ftz gene are localized in stripes corresponding to even numbered parasegments. The expressed regions correspond to the regions that are missing in the ftz mutant of the previous figure. Photograph kindly provided by Walter Gehring. |
The ftz mRNA is present from early blastoderm to gastrula stages of development. Figure 29.26 shows the locations of the transcripts, visualized in situ at blastoderm in wild type. The gene is expressed in 7 stripes, each 3 V4 cells wide, running across the embryo. As shown previously in Figure 29.24, the stripes correspond to even-numbered parasegments (4 = T1P/T2A, 6= T3P/A1A, 8 = A2P/A3A, etc.).
This pattern suggests a function for the ftz gene: it must be expressed at blastoderm for the structures that will be descended from the even-numbered parasegments to develop. Mutants in which ftz is defective lack these parasegments because the gene product is absent during the period when they must be formed. In other words, expression of ftz is required for survival of the cells in the regions in which it is expressed.
The expression of ftz is an example of the general rule that the stripes in which a pair-rule gene is expressed correspond to the regions that are missing from the embryo when the gene is mutated. Compartments are therefore determined by the pattern of expression of segmentation genes. The width of the stripe in which a gene is expressed corresponds to the size of the segmental unit that it affects. Different mechanisms are used to specify the expression patterns of different pair-rule genes; we have the most information about ftz and eve.
In the early embryo, ftz is uniformly expressed. If protein synthesis is blocked before the stripes develop, the embryo retains the initial pattern. So the development of stripes depends on the specific degradation of ftz RNA in the regions between the bands and at the anterior and posterior ends of the embryo. Once the stripes have developed, transcription of ftz ceases in the interbands and at the ends of the embryo. The specificity of transcription depends on regions upstream of the ftz promoter, and also on the function of several other segmentation genes. The transcription of ftz responds to other pair-rule genes (and perhaps gap genes) through elements that act on all stripes.
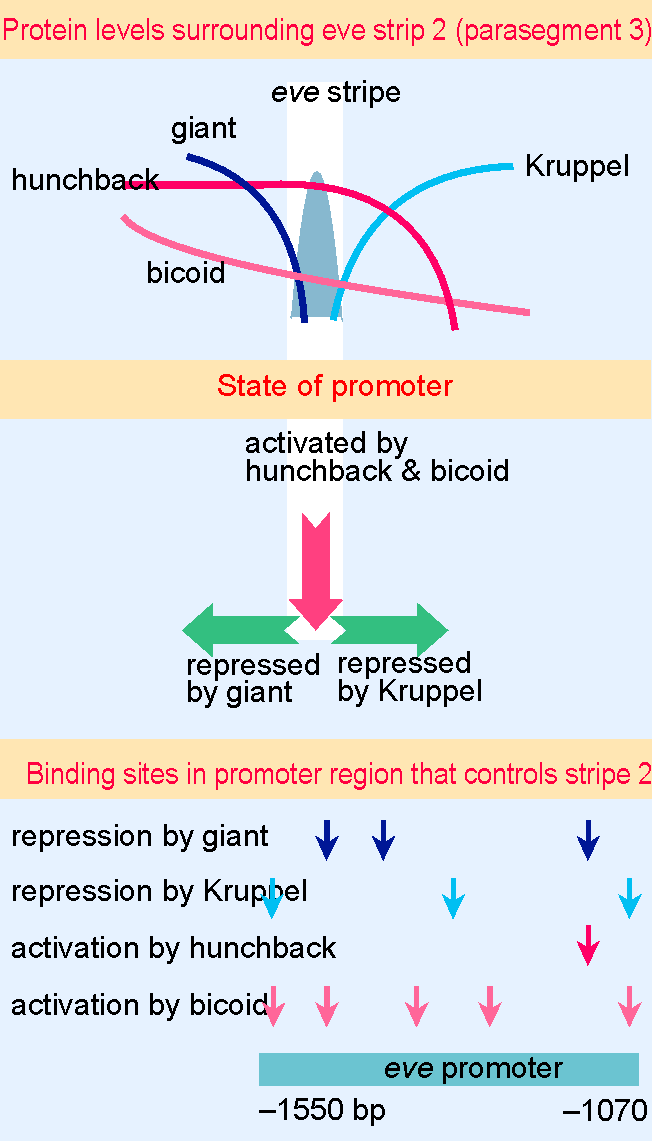 |
Figure 29.27 The eve stripe in parasegment 3 is activated by hunchback and bicoid. Repression by giant sets the anterior boundary; repression by Kruppel sets the posterior boundary. Mulltiple binding sites for these proteins in a 480 bp region of the promoter control expression of the gene. |
The expression pattern of eve is complementary to ftz, but has a different basis: it is controlled separately in each stripe. A detailed reconstruction using subregions of the eve promoter shows that the information for localization in each stripe is coded in a separate part of the promoter; the promoter can be divided into regions that correspond to the local levels of gap gene products in particular parasegments. For example, the promoter region that is responsible for eve expression in parasegment 3 has binding sites for the gap proteins bicoid, hunchback, giant, and Kruppel. Figure 29.27 shows that this part of the promoter extends for 480 bp. It works in the following way. eve transcription is activated by hunchback and bicoid. The two boundaries are determined because the promoter is repressed by giant on the anterior side and by Kruppel on the posterior side (see also Figure 29.24). Other parts of the promoter respond to the protein levels in other parts of the embryo. So the different stripes of the primary pair-rule gene products are regulated by separate pathways, each of which is susceptible to activation by a particular combination of gap gene products and other regulators.
This illustrates in miniature the principle that combinations of proteins control gene expression in local areas. The general principle is that generally distributed proteins (such as bicoid or hunchback) are needed for activation, whereas the borders are formed by selective repression (by giant and Kruppel in this particular example). We should emphasize that the hierarchy of gene control is not exclusively restricted to interactions between successive stages of control (maternal gap pair-rule). For example, the involvement of bicoid protein in regulating eve transcription in parasegment 3 shows that a maternal gene may have a direct effect on a pair-rule gene.
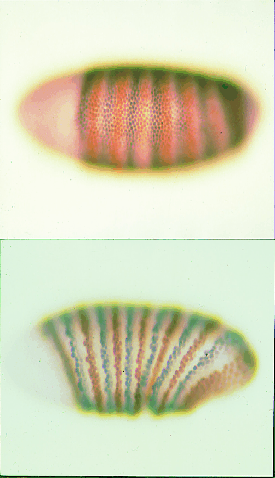 |
Figure 29.28 Simultaneous staining for ftz (brown) and eve (grey) shows that they are first expressed as broad alternating stripes at the time of blastoderm (upper), but narrow during the next 1 hour of development (lower). Photographs kindly provided by Peter Lawrence. |
The stripes of eve and ftz are fuzzy to begin with, and become sharper as development proceeds, corresponding to more finely defined units. Figure 29.28 shows an example of an embryo simultaneously stained for expression of ftz and eve. Initially there is a series of alternating fuzzy stripes, but the stripes narrow from the posterior margin and sharpen on the anterior side as they intensify during development. This may depend on an autoregulatory loop, in which the expression of the gene is regulated by its own product.
The pair-rule genes control the expression of the segment polarity genes, which are expressed in 14 stripes. Each stripe identifies a segment. The compartmental pattern in which segment polarity genes are expressed is exceedingly precise. Perhaps the ultimate demonstration of precision is provided by the pattern gene engrailed. The function coded by engrailed is needed in all segments and is concerned with the distinction between the A and P compartments. engrailed is expressed in every P compartment, but not in A compartments. Mutants in this gene do not distinguish between anterior and posterior compartments of the segments.
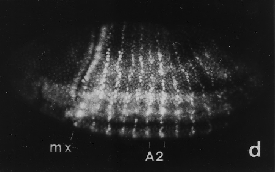 |
Figure 29.29 Engrailed protein is localized in nuclei and forms stripes as precisely delineated as 1 cell in width. Photograph kindly provided by Patrick O'Farrell. |
Antibodies against the protein coded by engrailed react against the nucleus of cells expressing it. The regions in which engrailed is expressed form a pattern of stripes. When the stripes of engrailed protein first become apparent, they are only one cell wide. Figure 29.29 shows the pattern at a stage when each segment has a stripe just 1 cell in width, with the stripe beginning to widen into several cells.
Actually, the pattern of stripes becomes established over a 30 minute period, moving along the embryo from anterior to posterior. Initially one stripe is apparent; then every other segment has a stripe; and finally the complete pattern has a stripe 3 V4 cells wide corresponding to the P compartment of every segment.
The expression of engrailed is of particular importance, because it defines the boundaries of the actual compartments from which adult structures will be derived. The initial 1-cell-wide stripes of engrailed protein form at the anterior boundaries of both the ftz and eve stripes, and delineate what will become the anterior boundary of every P compartment. Why is engrailed initially transcribed exclusively in this anterior edge, within the broader stripes of ftz and eve expression? This question is a specific example of a more general question: how can a broad stripe be subdivided into more restricted, narrower stripes? We can consider two general types of model:
- A combinatorial model supposes that different genes are expressed in overlapping patterns of stripes. A pattern of stripes develops for each of the pair-rule genes. The different pair-rule gene stripes overlap, because they are out of phase with one another. As a result of these patterns, different cells in the cellular blastoderm express different combinations of pair-rule genes. Each compartment is defined by the particular combination of the genes that are expressed, and these combinations determine the responses of the cells at next stage of development. In other words, the segmentation genes are controlled by the pair-rule genes in the same general manner that the pair-rule genes are controlled by the gap genes.
- A boundary model supposes that a compartment is defined by the striped pattern of expression, but that interactions involving cell-cell communication at the boundaries cause subdivisions to arise within the compartment. In the case of engrailed, we would suppose that some unique event is triggered by the juxtaposition of cells possessing ftz (or eve) with cells that do not, and this is necessary to trigger engrailed expression.
Each of the 14 segments is subdivided further by functions of the segment polarity genes. The type of subdivision imposed by a segment polarity gene is repeated in every segment. For example, engrailed distinguishes the A and P compartments. engrailed is a transcription factor, but other segment polarity genes have different types of functions. wingless codes for a protein that is secreted and taken up by nearby cells, so this allows cell-cell interactions to play a role. wingless is initially expressed in a row of cells immediately adjacent to the anterior side of the cells expressing engrailed; thus wingless comes to identify the posterior boundary of the preceding parasegment. The initial expression of engrailed in response to ftz and eve is shortly replaced by an autoregulatory loop in which secretion of wingless protein from the adjacent cells is needed for expression of engrailed; and expression of engrailed is needed for expression of wingless. This keeps the boundary sharp. We do not yet have a good understanding of how the other segment polarity genes function to delineate the segments or parts of segments. However, their products include secreted proteins, transmembrane proteins, kinases, cytoskeletal proteins, as well as transcription factors, suggesting that at this stage cell-cell communications become important.
| Reviews | |
| Ingham, P. W. and Martinez-Arias, A. (1992). Boundaries and fields in early embryos. Cell 68, 221-235. | |
| Scott, M. P. and Carroll, S. B. (1987). The segmentation and homeotic gene network in early Drosophila development. Cell 51, 689-698. | |
| Research | |
| Nusslein-Vollhard, C. and Wieschaus, E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795-801. | |