7. RNA editing utilizes information from several sources
23.6 RNA can have ribonuclease activities |
| Key terms defined in this section |
| Viroid is a small infectious nucleic acid that does not have a protein coat. |
One of the first demonstrations of the capabilities of RNA was provided by the dissection of ribonuclease P, an E. coli tRNA-processing endonuclease. Ribonuclease P can be dissociated into its two components, the 375 base RNA and the 20 kD polypeptide. Under the conditions initially used to characterize the enzyme activity in vitro, both components were necessary to cleave the tRNA substrate.
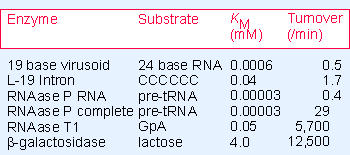 |
Figure 23.10 Reactions catalyzed by RNA have the same features as those catalyzed by proteins, although the rate is slower. The KM gives the concentration of substrate required for half-maximum velocity; this is an inverse measure of the affinity of the enzyme for substrate. The turnover number gives the number of substrate molecules transformed in unit time by a single catalytic site. |
But a change in ionic conditions, an increase in the concentration of Mg2+, renders the protein component superfluous. The RNA alone can catalyze the reaction! Analyzing the results as though the RNA were an enzyme, each "enzyme" catalyzes the cleavage of multiple substrates. Although the catalytic activity resides in the RNA, the protein component greatly increases the speed of the reaction, as seen in the increase in turnover number (see Figure 23.10).
Because mutations in either the gene for the RNA or the gene for protein can inactivate RNAase P in vivo, we know that both components are necessary for natural enzyme activity. Originally it had been assumed that the protein provided the catalytic activity, while the RNA filled some subsidiary role, for example, assisting in the binding of substrate (it has some short sequences complementary to exposed regions of tRNA). But these roles are reversed!
The catalytic activity of RNAase P requires RNA to function as a catalyst with an external substrate. Another example of the ability of RNA to function as an endonuclease is provided by some small plant RNAs (~350 bases) that undertake a self-cleavage reaction. As with the case of the Tetrahymena group I intron, however, it is possible to engineer constructs that can function on external substrates (Guerrier-Takada et al., 1983).
These small plant RNAs fall into two general groups: viroids and virusoids. The viroids are infectious RNA molecules that function independently, without enacapsidation by any protein coat. The virusoids are similar in organization, but are encapsidated by plant viruses, being packaged together with a viral genome. The virusoids cannot replicate independently, but require assistance from the virus. The virusoids are sometimes called satellite RNAs.
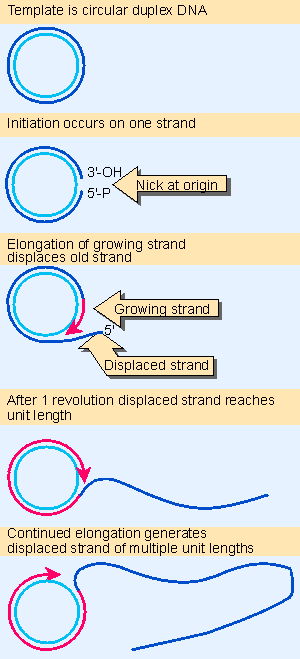 |
Figure 12.16 The rolling circle generates a multimeric single-stranded tail. Animated figure |
Viroids and virusoids both replicate via rolling circles (see Figure 12.16). The strand of RNA that is packaged into the virus is called the plus strand. The complementary strand, generated during replication of the RNA, is called the minus strand. Multimers of both plus and minus strands are found. Both types of monomer are generated by cleaving the tail of a rolling circle; circular plus strand monomers are generated by ligating the ends of the linear monomer.
Both plus and minus strands of viroids and virusoids undergo self-cleavage in vitro. The cleavage reaction is promoted by divalent metal cations; it generates 5′ VOH and 2′ V3′-cyclic phosphodiester termini. Some of the RNAs cleave in vitro under physiological conditions. Others do so only after a cycle of heating and cooling; this suggests that the isolated RNA has an inappropriate conformation, but can generate an active conformation when it is denatured and renatured.
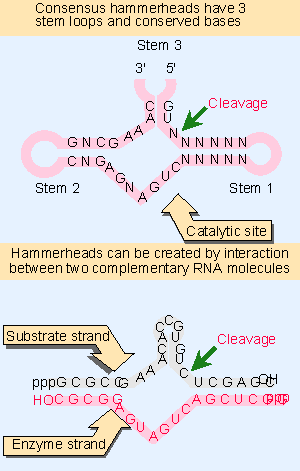 |
Figure 23.13 Self-cleavage sites of viroids and virusoids have a consensus sequence and form a hammerhead secondary structure by intramolecular pairing. Hammerheads can also be generated by pairing between a substrate strand and an "enzyme" strand. |
The viroids and virusoids that undergo self-cleavage form a "hammerhead" secondary structure at the cleavage site, as drawn in the upper part of Figure 23.13. The sequence of this structure is sufficient for cleavage. When the surrounding sequences are deleted, the need for a heating-cooling cycle is obviated, and the small RNA self-cleaves spontaneously. This suggests that the sequences beyond the hammerhead usually interfere with its formation.
The active site is a sequence of only 58 nucleotides. The hammerhead contains three stem-loop regions whose position and size are constant, and 13 conserved nucleotides, mostly in the regions connecting the center of the structure. The conserved bases and duplex stems generate an RNA with the intrinsic ability to cleave (Forster and Symons, 1987).
An active hammerhead can also be generated by pairing an RNA representing one side of the structure with an RNA representing the other side. The lower part of Figure 23.13 shows an example of a hammerhead generated by hybridizing a 19 base molecule with a 24 base molecule. The hybrid mimics the hammerhead structure, with the omission of loops I and III. When the 19 base RNA is added to the 24 base RNA, cleavage occurs at the appropriate position in the hammerhead.
We may regard the top (24 base) strand of this hybrid as comprising the "substrate," and the bottom (19 base) strand as comprising the "enzyme." When the 19 base RNA is mixed with an excess of the 24 base RNA, multiple copies of the 24 base RNA are cleaved. This suggests that there is a cycle of 19 base V24 base pairing, cleavage, dissociation of the cleaved fragments from the 19 base RNA, and pairing of the 19 base RNA with a new 24 base substrate. The 19 base RNA is therefore a ribozyme with endonuclease activity. The parameters of the reaction are similar to those of other RNA-catalyzed reactions (see Figure 23.10; for review see Symons, 1992).
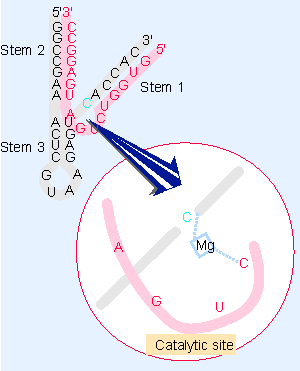 |
Figure 23.14 A hammerhead ribozyme forms a V-shaped tertiary structure in which stem 2 is stacked upon stem 3. The catalytic center lies between stem 2/3 and stem 1. It contains a magnesium ion that initiates the hydrolytic reaction. |
The crystal structure of a hammerhead shows that it forms a compact V-shape, in which the catalytic center lies in a turn, as indicated diagrammatically in Figure 23.14. An Mg2+ ion located in the catalytic site plays a crucial role in the reaction. It is positioned by the target cytidine and by the cytidine at the base of stem 1; it may also be connected to the adjacent uridine. It extracts a proton from the 2′-OH of the target cytidine, and then directly attacks the labile phosphodiester bond. Mutations in the hammerhead sequence that affect the transition state of the cleavage reaction occur in both the active site and other locations, suggesting that there may be a substantial rearrangement of structure prior to cleavage (Scott et al., 1995).
It is possible to design enzyme-substrate combinations that can form hammerhead structures, and these have been used to demonstrate that introduction of the appropriate RNA molecules into a cell can allow the enzymatic reaction to occur in vivo. A ribozyme designed in this way essentially provides a highly specific restriction-like activity directed against an RNA target. By placing the ribozyme under control of a regulated promoter, it can be used in the same way as (for example) antisense constructs specifically to turn off expression of a target gene under defined circumstances.
| Reviews | |
| Symons, R. H. (1992). Small catalytic RNAs. Ann. Rev. Biochem 61, 641-71. | |
| Research | |
| Forster, A. C. and Symons, R. H. (1987). Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell 50, 9-16. | |
| Guerrier-Takada, C. et al. (1983). The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849-857. | |
| Scott, W. G., Finch, J. T., and Klug, A. (1995). The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell 81, 991-1002. | |