6. Alternative splicing involves differential use of splice junctions
22.5 Group II introns autosplice via lariat formation |
Introns in protein-coding genes (in fact, in all genes except nuclear tRNA-coding genes) can be divided into three general classes. Nuclear pre-mRNA introns have in common only the possession of the GT...AG dinucleotides at the 5′ and 3′ ends. Group I and group II introns are found in organelles and in bacteria. (Group I introns are found also in the nucleus in lower eukaryotes.) Group I and group II introns are classified according to their internal organization. Each can be folded into a typical type of secondary structure.
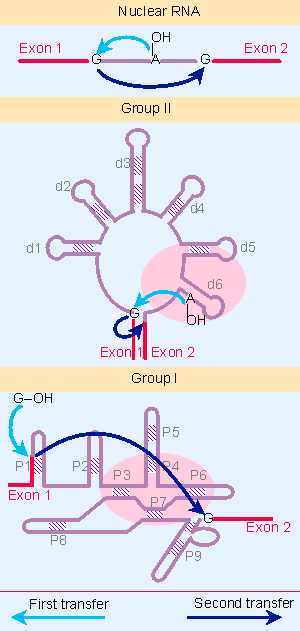 |
Figure 22.15 Three classes of splicing reactions proceed by two transesterifications. First, a free OH group attacks the exon 1 - intron junction. Second, the OH created at the end of exon 1 attacks the intron - exon 2 junction. |
 |
Figure 22.6 Splicing occurs in two stages, in which the 5 F exon is separated and then is joined to the 3 F exon. |
Figure 22.15 shows that three classes of introns are excised by a two successive transesterifications (shown previously for nuclear introns in Figure 22.6). In the first reaction, the 5′ exon-intron junction is attacked by a free hydroxyl group (provided by an internal 2′ VOH position in nuclear and group II introns, and by a free guanine nucleotide in group I introns). In the second reaction, the free 3′ VOH at the end of the released exon in turn attacks the 3′ intron-exon junction.
Group I introns are more common than group II introns. There is little relationship between the two classes, but they share the striking property that the RNA can perform the splicing reaction in vitro by itself, without requiring enzymatic activities provided by proteins; however, proteins are almost certainly required in vivo to assist with folding. We discuss the catalytic reaction of group I introns in 23 Catalytic RNA, but now we examine the parallels between group II introns and nuclear splicing.
 |
Figure 22.16 Splicing releases mitochondrial group II introns in the form of stable lariats. Photograph kindly provided by Leslie Grivell and Annika Arnberg. |
Group II mitochondrial introns have splice sites that resemble nuclear splice sites. They are excised by the same mechanism as nuclear pre-mRNAs, via a lariat that is held together by a 5′ V2′ bond. An example of a lariat produced by splicing a group II intron is shown in Figure 22.16. When an isolated group II RNA is incubated in vitro in the absence of additional components, it is able to perform the splicing reaction. This means that the two transesterification reactions shown in Figure 22.15 can be performed by the group II intron RNA sequence itself. Because the number of phosphodiester bonds is conserved in the reaction, an external supply of energy is not required; this could have been an important feature in the evolution of splicing (for review see Michel and Ferat, 1995).
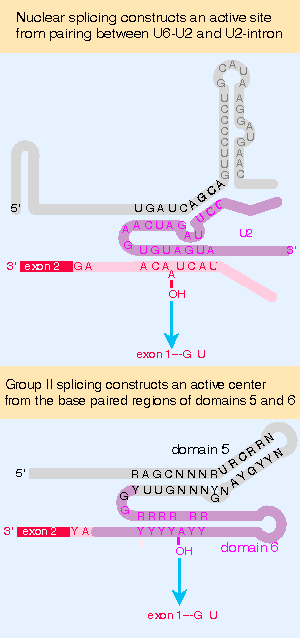 |
Figure 22.17 Nuclear splicing and group II splicing involve the formation of similar secondary structures. The sequences are more specific in nuclear splicing; group II splicing uses positions that may be occupied by either purine (R) or either pyrimidine (Y). |
A group II intron forms into a secondary structure that involves closely juxtaposing two of its domains. Domain 5 is separated by 2 bases from domain 6, which contains an A residue that donates the 2′ VOH group for the first transesterification. This constitutes a catalytic domain in the RNA. Figure 22.17 compares this secondary structure with the structure formed by the combination of U6 with U2 and of U2 with the branch site. The similarity suggests that U6 may have a catalytic role.
The features of group II splicing suggest that splicing evolved from an autocatalytic reaction undertaken by an individual RNA molecule, in which it accomplished a controlled deletion of an internal sequence. Probably such a reaction requires the RNA to fold into a specific conformation, or series of conformations, and would occur exclusively in cis conformation.
The ability of group II introns to remove themselves by an autocatalytic splicing event stands in great contrast to the requirement of nuclear introns for a complex splicing apparatus. We may regard the snRNAs of the spliceosome as compensating for the lack of sequence information in the intron, and providing the information required to form particular structures in RNA. The functions of the snRNAs may have evolved from the original autocatalytic system. These snRNAs act in trans upon the substrate pre-mRNA; we might imagine that the ability of U1 to pair with the 5′ splice site, or of U2 to pair with the branch sequence, replaced a similar reaction that required the relevant sequence to be carried by the intron. So the snRNAs may undergo reactions with the pre-mRNA substrate and with one another that have substituted for the series of conformational changes that occur in RNAs that splice by group II mechanisms. In effect, these changes have relieved the substrate pre-mRNA of the obligation to carry the sequences needed to sponsor the reaction. Of course, as the splicing apparatus has become more complex (and as the number of potential substrates has increased) some of the reactions that used to be catalyzed by RNA have been taken over by proteins.
| Reviews | |
| Michel, F. and Ferat, J.-L. (1995). Structure and activities of group II introns. Ann. Rev. Biochem 64, 435-461. | |
- Chapter VI Web Site Quality and Usability in E-Commerce
- Chapter IX Extrinsic Plus Intrinsic Human Factors Influencing the Web Usage
- Chapter XII Web Design and E-Commerce
- Chapter XIII Shopping Agent Web Sites: A Comparative Shopping Environment
- Chapter XVIII Web Systems Design, Litigation, and Online Consumer Behavior