7. cis-splicing and trans-splicing reactions
22.6 Alternative splicing involves differential use of splice junctions |
The majority of interrupted genes are transcribed into an RNA that gives rise to a single type of spliced mRNA: in these cases, there is no variation in assignment of exons and introns. But the RNAs of some genes follow patterns of alternative splicing, when a single gene gives rise to more than one mRNA sequence. In some cases, the ultimate pattern of expression is dictated by the primary transcript, because the use of different startpoints or termination sequences alters the pattern of splicing. In other cases, a single primary transcript is spliced in more than one way, and internal exons are substituted, added, or deleted. In some cases, the multiple products all are made in the same cell, but in others the process is regulated so that particular splicing patterns occur only under particular conditions (for review see Green, 1991).
One of the most pressing questions in splicing is to determine what controls the use of such alternative pathways. Proteins that intervene to bias the use of alternative splice sites have been identified in two ways. In some mammalian systems, it has been possible to characterize alternative splicing in vitro, and to identify proteins that are required for the process. In D. melanogaster, aberrations in alternative splicing may be caused either by mutations in the genes that are alternatively spliced or in the genes whose products are necessary for the reaction.
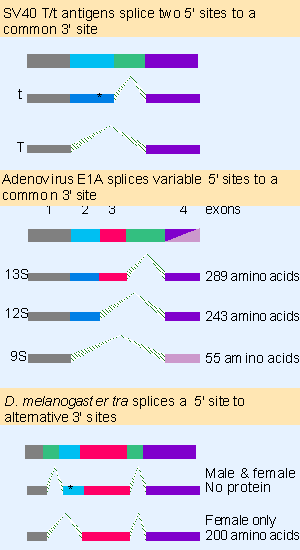 |
Figure 22.18 Alternative forms of splicing may generate a variety of protein products from an individual gene. Changing the splice sites may introduce termination codons (shown by asterisks) or change reading frames. Multiple figure |
Figure 22.18 shows examples in which one splice site remains constant, but the other varies. The large T/ small t antigens of SV40 and the products of the adenovirus E1A region are generated by connecting a varying 5′ site to a constant 3′ site. In the case of the T/t antigens, the 5′ site used for T antigen removes a termination codon that is present in the t antigen mRNA, so that T antigen is larger than t antigen. In the case of the E1A transcripts, one of the 5′ sites connects to the last exon in a different reading frame, again making a significant change in the C-terminal part of the protein. In these examples, all the relevant splicing events take place in every cell in which the gene is expressed, so all the protein products are made.
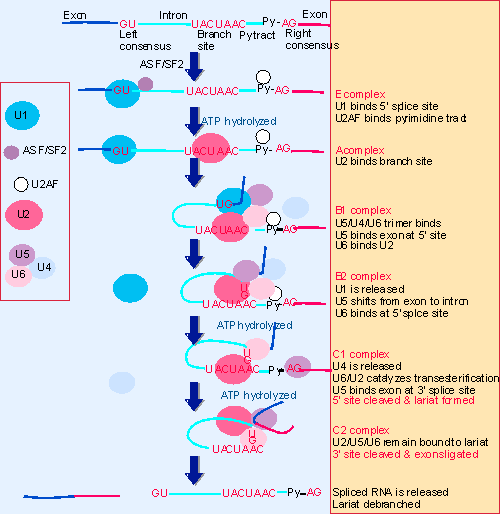 |
Figure 22.10 The splicing reaction proceeds through discrete stages in which spliceosome formation involves the interaction of components that recognize the consensus sequences. |
There are differences in the ratios of T/t antigens in different cell types. Extracts from cells that produce relatively more small t antigen during an infection also produce more RNA with its characteristic splicing pattern in an in vitro system. A protein extracted from these cells can cause preferential production of small t RNA in extracts from other cell types. This protein, which was called ASF (alternative splicing factor), turns out to be the same as the splicing factor SF2, which is required for early steps in spliceosome assembly and for the first cleavage-ligation reaction (see Figure 22.10). ASF/SF2 is an RNA-binding protein in the SR family. When a pre-mRNA has more than one 5′ splice site preceding a single 3′ splice site, increased concentrations of ASF/SF2 promote use of the 5′ site nearest to the 3′ site at the expense of the other site. This effect of ASF/SF2 can be counteracted by another splicing factor, SF5. The exact molecular roles of these factors are not yet known, but we see in general terms that alternative splicing involving different 5′ sites may be influenced by proteins involved in spliceosome assembly.
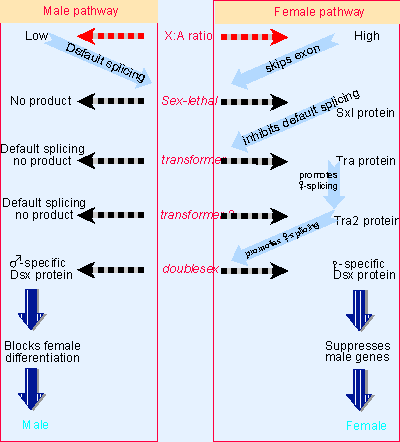 |
Figure 22.19 Sex determination in D. melanogaster involves a pathway in which different splicing events occur in females. Blocks at any stage of the pathway result in male development. |
The pathway of sex determination in D. melanogaster involves interactions between a series of genes in which alternative splicing events distinguish male and female. The pathway takes the form illustrated in Figure 22.19, in which the ratio of X chromosomes to autosomes determines the expression of sxl, and changes in expression are passed sequentially through the other genes to dsx, the last in the pathway.
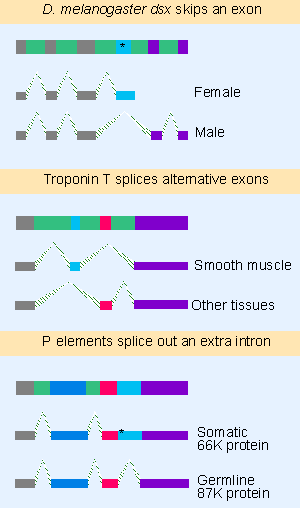 |
Figure 22.20 Alternative splicing events that involve both sites may cause exons to be added or substituted. |
The pathway starts with sex-specific splicing of sxl, following the same pattern that is shown below in Figure 22.20 for dsx. Exon 3 of the sxl gene contains a termination codon that prevents synthesis of functional protein. This exon is included in the mRNA produced in males, but is skipped in females. As a result, only females produce Sxl protein. The protein has a concentration of basic amino acids that resembles other RNA-binding proteins.
The presence of Sxl protein changes the splicing of the transformer (tra) gene. Figure 22.18 shows that this involves splicing a constant 5′ site to alternative 3′ sites. One splicing pattern occurs in both males and females, and results in an RNA that has an early termination codon. The presence of Sxl protein inhibits usage of the normal 3′ splice site; when this site is skipped, the next 3′ site is used, leaving out a whole exon. This generates a female-specific mRNA that codes for a protein.
So tra produces a protein only in females; this protein is a splicing regulator. Similarly tra2 is productively expressed in females but not in males. The Tra and Tra2 proteins are SR splicing factors that act directly upon the target transcripts. Tra and Tra2 cooperate (in females) to affect the splicing of dsx.
Figure 22.20 shows examples of cases in which splice sites are used to add or to substitute exons or introns, again with the consequence that different protein products are generated. In the doublesex (dsx) gene, females splice the 5′ site of intron 3 to the 3′ site of that intron; as a result translation terminates at the end of exon 4. Males splice the 5′ site of intron 3 directly to the 3′ site of intron 4, thus omitting exon 4 from the mRNA, and allowing translation to continue through exon 6. The result of the alternative splicing is that different proteins are produced in each sex: the male product blocks female sexual differentiation, while the female product represses expression of male-specific genes.
Sex determination therefore has a pleasing symmetry: the pathway starts with a female-specific splicing event that causes omission of an exon that has a termination codon, and ends with a female-specific splicing event that causes inclusion of an exon that has a termination codon. The events have different molecular bases. At the first control point, Sxl inhibits the default splicing pattern. At the last control point, Tra and Tra2 cooperate to promote the female-specific splice.
Alternative splicing of dsx RNA is controlled by competition between 3′ splice sites. dsx RNA has an element downstream of the leftmost 3′ splice site that is bound by Tra2; Tra and SR proteins associate with Tra2 at the site, which becomes an enhancer that assists binding of U2AF at the adjacent branch site. This commits the formation of the spliceosome to use this 3′ site in females rather than the alternative 3′ site. The proteins recognize the enhancer cooperatively, possibly relying on formation of some secondary structure as well as sequence per se.
The Tra and Tra2 proteins are not needed for normal splicing, because in their absence flies develop as normal males. As specific regulators, they need not necessarily participate in the mechanics of the splicing reaction; in this respect they differ from SF2, which is a factor required for general splicing, but can also influence choice of alternative splice sites.
Competition between 5′ splice sites also may be controlled by specific regulators. In one case, binding of SR proteins near the site favors its use, presumably by increasing the affinity for U1. This may identify a type of enhancer that functions on the 5′ site.
Alternative splicing also may be influenced by repression of one site. Exons 2 and 3 of the mouse troponin T gene are mutually exclusive; exon 2 is used in smooth muscle, but exon 3 is used in other tissues. Smooth muscle contains proteins that bind to repeated elements located on either side of exon 3, and which prevent use of the 3′ and 5′ sites that are needed to include it.
P elements of D. melanogaster show a tissue-specific splicing pattern. In somatic cells, there are two splicing events, but in germline an additional splicing event removes another intron. Because a termination codon lies in the germline-specific intron, a longer protein (with different properties) is produced in germline. We discuss the consequences for control of transposition in 15 Transposons, and note for now that the tissue specificity results from differences in the splicing apparatus.
The default splicing pathway when the RNA is subjected to a heterologous (human) splicing extract is the germline pattern, in which intron 3 is excised. But extracts of somatic cells of D. melanogaster contain a protein that inhibits excision of this intron. The protein binds to sequences in exon 3; if these sequences are deleted, the intron is excised. The function of the protein is therefore probably to repress association of the spliceosome with the 5′ site of intron 3.
| Reviews | |
| Green, M. R. (1991). Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Ann. Rev. Cell Biol. 7, 559-599. | |