11. The genetic code is triplet
1.11 The genetic code is triplet |
| Key terms defined in this section |
| Codon is a triplet of nucleotides that represents an amino acid or a termination signal. Frameshift mutation results from an insertion or deletion that changes the phase of triplets, so that all codons are misread after the site of mutation. Genetic code is the correspondence between triplets in DNA (or RNA) and amino acids in protein. Initiation codon is a special codon (usually AUG) used to start synthesis of a protein. ORF is an open reading frame; presumed likely to code for a protein.> Reading frame is one of three possible ways of reading a nucleotide sequence as a series of triplets. Suppressor (extragenic) is usually a gene coding a mutant tRNA that reads the mutated codon either in the sense of the original codon or to give an acceptable substitute for the original meaning. Termination codon is one of three (UAG, UAA, UGA) that causes protein synthesis to terminate. |
Each gene represents a particular polypeptide chain. The concept that each protein consists of a particular series of amino acids dates from Sanger’s characterization of insulin in the 1950s. The discovery that a gene consists of DNA faces us with the issue of how a sequence of nucleotides in DNA represents a sequence of amino acids in protein.
A crucial feature of the general structure of DNA is that it is independent of the particular sequence of its component nucleotides. The sequence of nucleotides in DNA is important not because of its structure per se, but because it codes for the sequence of amino acids that constitutes the corresponding polypeptide. The relationship between a sequence of DNA and the sequence of the corresponding protein is called the genetic code.
The structure and/or enzymatic activity of each protein follows from its primary sequence of amino acids. By determining the sequence of amino acids in each protein, the gene is able to carry all the information needed to specify an active polypeptide chain. In this way, a single type of structure Xthe gene Xis able to represent itself in innumerable polypeptide forms.
Together the various protein products of a cell undertake the catalytic and structural activities that are responsible for establishing its phenotype. Of course, in addition to sequences that code for proteins, DNA also contains certain sequences whose function is to be recognized by regulator molecules, usually proteins. Here the function of the DNA is determined by its sequence directly, not via any intermediary code. Both types of region, genes expressed as proteins and sequences recognized as such, constitute genetic information.
The genetic code is deciphered by a complex apparatus that interprets the nucleic acid sequence. This apparatus is essential if the information carried in DNA is to have meaning. In any given region, only one of the two strands of DNA codes for protein, so we write the genetic code as a sequence of bases (rather than base pairs).
The genetic code is read in groups of three nucleotides, each group representing one amino acid. Each trinucleotide sequence is called a codon. A gene includes a series of codons that is read sequentially from a starting point at one end to a termination point at the other end. Written in the conventional 5′→3′ direction, the nucleotide sequence of the DNA strand that codes for protein corresponds to the amino acid sequence of the protein written in the direction from N-terminus to C-terminus.
The genetic code is read in nonoverlapping triplets from a fixed starting point:
- Nonoverlapping implies that each codon consists of three nucleotides and that successive codons are represented by successive trinucleotides.
- The use of a fixed starting point means that assembly of a protein must start at one end and work to the other, so that different parts of the coding sequence cannot be read independently.
The nature of the code predicts that two types of mutations will have different effects. If a particular sequence is read sequentially, such as:
UUU AAA GGG CCC (codons)
aa1 aa2 aa3 aa4 (amino acids)
then a point mutation will affect only one amino acid. For example, the substitution of an A by some other base (X) causes αα2 to be replaced by αα5:
UUU AAX GGG CCC
aa1 aa5 aa3 aa4
because only the second codon has been changed.
But a mutation that inserts or deletes a single base will change the reading frame for the entire subsequent sequence. A change of this sort is called a frameshift. An insertion might take the form:
UUU AAX AGG GCC C
aa1 aa5 aa6 aa7
Because the new sequence of triplets is completely different from the old one, the entire amino acid sequence of the protein is altered beyond the site of mutation. So the function of the protein is likely to be lost completely.
Frameshift mutations are induced by the acridines, compounds that bind to DNA and distort the structure of the double helix, causing additional bases to be incorporated or omitted during replication. Each mutagenic event sponsored by an acridine results in the addition or removal of a single base pair (for review see Roth, 1974).
If an acridine mutant is produced by, say, addition of a nucleotide, it should revert to wild type by deletion of the nucleotide. But reversion can also be caused by deletion of a different base, at a site close to the first. Combinations of such mutations provided revealing evidence about the nature of the genetic code.
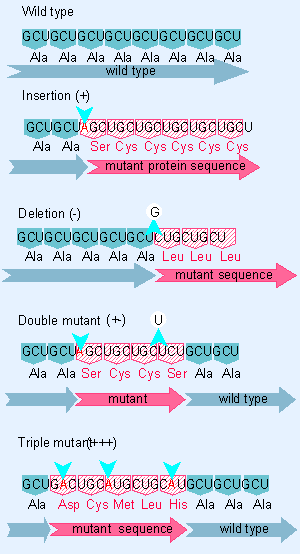 |
Figure 1.25 Frameshift mutations show that the genetic code is read in triplets from a fixed starting point. |
Figure 1.25 illustrates the properties of frameshift mutations. An insertion or a deletion changes the entire protein sequence following the site of mutation. But the combination of an insertion and a deletion causes the code to be read in the incorrect frame only between the two sites of mutation; correct reading resumes after the second site.
Genetic analysis of acridine mutations in the rII region of the phage T6 in 1961 showed that all the mutations could be classified into one of two sets, described as (+) and ( V). Either type of mutation by itself causes a frameshift, the (+) type by virtue of a base addition, the ( V) type by virtue of a base deletion. Double mutant combinations of the types (+ +) and ( V V) continue to show mutant behavior. But combinations of the types (+ V) or ( V +) suppress one another, giving rise to a description in which one mutation is described as a suppressor of the other. (In the context of this work, "suppressor" is used in an unusual sense, because the second mutation is in the same gene as the first.)
These results show that the genetic code must be read as a sequence that is fixed by the starting point, so additions or deletions compensate for each other, whereas double additions or double deletions remain mutant. But this does not reveal how many nucleotides make up each codon.
When triple mutants are constructed, only (+ + +) and ( V V V ) combinations show the wild phenotype, while other combinations remain mutant. If we take three additions or three deletions to correspond respectively to the addition or omission overall of a single amino acid, this implies that the code is read in triplets. An incorrect amino acid sequence is found between the two outside sites of mutation, and the sequence on either side remains wild type, as indicated in Figure 1.25 (Benzer and Champe, 1961; Crick et al., 1961).
If the genetic code is read in nonoverlapping triplets, there are three possible ways of translating any nucleotide sequence into protein, depending on the starting point. These called reading frames. For the sequence
A C G A C G A C G A C G A C G A C G
the three possible reading frames are
ACG ACG ACG ACG ACG ACG ACG
CGA CGA CGA CGA CGA CGA CGA
GAC GAC GAC GAC GAC GAC GAC
A reading frame that consists exclusively of triplets that represent amino acids is called an open reading frame or ORF. A sequence that is translated into protein has a reading frame that starts with a special initiation codon (AUG) and that extends through a series of triplets representing amino acids until it ends at one of three types of termination codon (see 5 Messenger RNA).
A reading frame that cannot be read into protein because termination codons occur frequently is said to be blocked. If a sequence is blocked in all three reading frames, it cannot have the function of coding for protein.
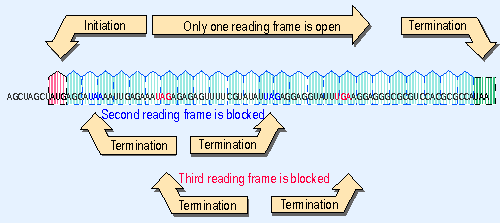 |
Figure 1.26 An open reading frame starts with AUG and continues in triplets to a termination codon. Blocked reading frames may be interrupted frequently by termination codons. |
When the sequence of a DNA region of unknown function is obtained, each possible reading frame is analyzed to determine whether it is open or blocked. Usually no more than one of the three possible frames of reading is open in any single stretch of DNA. Figure 1.26 shows an example of a sequence that can be read in only one reading frame, because the alternative reading frames are blocked by frequent termination codons. A long open reading frame is unlikely to exist by chance; if it were not translated into protein, there would have been no selective pressure to prevent the accumulation of termination codons. So the identification of a lengthy open reading frame is taken to be prima facie evidence that the sequence is translated into protein in that frame. An open reading frame (ORF) for which no protein product has been identified is sometimes called an unidentified reading frame (URF).
| Reviews | |
| Roth, J. R. (1974). Frameshift mutations. Ann. Rev. Genet. 8, 319-346. | |
| Research | |
| Benzer, S. and Champe, S. P. (1961). Ambivalent rII mutants of phage T4. Proc. Nat. Acad. Sci. USA 47, 403-416. | |
| Crick, F. H. C., Barnett, L., Brenner, S., and Watts-Tobin, R. J. (1961). General nature of the genetic code for proteins. Nature 192, 1227-1232. | |