1. Introduction
10.1 Introduction |
| Key terms defined in this section |
| Operator is the site on DNA at which a repressor protein binds to prevent transcription from initiating at the adjacent promoter. Repressor protein binds to operator on DNA or RNA to prevent transcription or translation, respectively. Structural gene codes for any RNA or protein product other than a regulator. |
Gene expression can be controlled at any of several stages, which we divide broadly into transcription, processing, and translation:
- Transcription often is controlled at the stage of initiation. Transcription is not usually controlled at elongation, but may be controlled at termination to prevent transcription from proceeding past a terminator to the gene(s) beyond.
- In eukaryotic cells, processing of the RNA product may be regulated at the stages of modification, splicing, transport, or stability. In bacteria, an mRNA is in principle available for translation as soon as it is synthesized, and these stages of control are not available.
- Translation may be regulated, usually at the stages of initiation and termination (like transcription). Regulation of initiation is formally analogous to the regulation of transcription: the circuitry can be drawn in similar terms for regulating initiation of transcription on DNA or initiation of translation on RNA.
The basic concept for how transcription is controlled in bacteria was provided by the classic formulation of the model for control of gene expression by Jacob and Monod in 1961 (Jacob and Monod, 1961). They distinguished between two types of sequences in DNA: sequences that code for trans-acting products; and cis-acting sequences that function exclusively within the DNA. Gene activity is regulated by the specific interactions of the trans-acting products (usually proteins) with the cis-acting sequences (usually sites in DNA). In more formal terms:
- A gene is a sequence of DNA that codes for a diffusible product. This product may be protein (as in the case of the majority of genes) or may be RNA (as in the case of genes that code for tRNA and rRNA). The crucial feature is that the product diffuses away from its site of synthesis to act elsewhere. Any gene product that is free to diffuse to find its target is described as trans-acting.
- The description cis-acting applies to any sequence of DNA that is not converted into any other form, but that functions exclusively as a DNA sequence in situ, affecting only the DNA to which it is physically linked. (In some cases, a cis-acting sequence functions in an RNA rather than in a DNA molecule.)
To help distinguish between the components of regulatory circuits and the genes that they regulate, we sometimes use the terms structural gene and regulator gene. A structural gene is simply any gene that codes for a protein (or RNA) product. Structural genes represent an enormous variety of protein structures and functions, including structural proteins, enzymes with catalytic activities, and regulatory proteins. A regulator gene simply describes a gene that codes for a protein involved in regulating the expression of other genes.
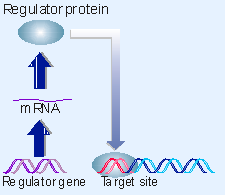 |
Figure 10.1 A regulator gene codes for a protein that acts at a target site on DNA. |
The crux of the regulatory model is illustrated in Figure 10.1: a regulator gene codes for a protein that controls transcription by binding to particular site(s) on DNA. This interaction can regulate a target gene in either a positive manner (the interaction turns the gene on) or in a negative manner (the interaction turns the gene off). The sites on DNA are usually (but not exclusively) located just upstream of the target gene.
The sequences that mark the beginning and end of the transcription unit, the promoter and terminator, are examples of cis-acting sites. A promoter serves to initiate transcription only of the gene or genes physically connected to it on the same stretch of DNA. In the same way, a terminator can terminate transcription only by an RNA polymerase that has traversed the preceding gene(s). In their simplest forms, promoters and terminators are cis-acting elements that are recognized by the same trans-acting species, that is, by RNA polymerase (although other factors also participate at each site).
Additional cis-acting regulatory sites are often juxtaposed to, or interspersed with, the promoter. A bacterial promoter may have one or more such sites located close by, that is, in the immediate vicinity of the startpoint. A eukaryotic promoter is likely to have a greater number of sites, spread out over a longer distance.
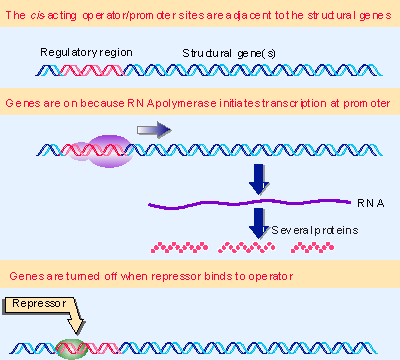 |
Figure 10.2 In negative control, a trans-acting repressor binds to the cis-acting operator to turn off transcription. In prokaryotes, multiple genes are controlled coordinately. |
A classic mode of control in bacteria is negative: a repressor protein prevents a gene from being expressed. Figure 10.2 shows that the "default state" for such a gene is to be expressed via the recognition of its promoter by RNA polymerase. Close to the promoter is another cis-acting site called the operator, which is the target for the repressor protein. When the repressor binds to the operator, RNA polymerase is prevented from initiating transcription, and gene expression is therefore turned off.
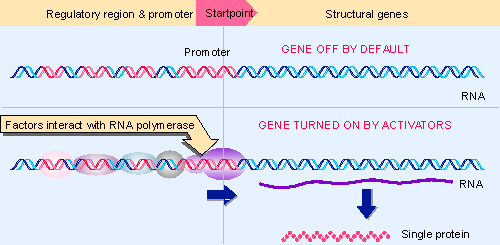 |
Figure 10.3 In positive control, trans-acting factors must bind to cis-acting sites in order for RNA polymerase to initiate transcription at the promoter. In a eukaryotic system, a structural gene is controlled individually.< |
An alternative mode of control is positive. This is used in bacteria (probably) with about equal frequency to negative control, and it is the most common mode of control in eukaryotes. A transcription factor is required to assist RNA polymerase in initiating at the promoter. Figure 10.3 shows that the typical default state of a eukaryotic gene is inactive: RNA polymerase cannot by itself initiate transcription at the promoter. Several trans-acting factors have target sites in the vicinity of the promoter, and binding of some or all of these factors enables RNA polymerase to initiate transcription.
The unifying theme is that regulatory proteins are trans-acting factors that recognize cis-acting elements (usually) upstream of the gene. The consequences of this recognition are to activate or to repress the gene, depending on the individual type of regulatory protein. A typical feature is that the protein functions by recognizing a very short sequence in DNA, usually <10 bp in length, although the protein actually binds over a somewhat greater distance of DNA. The bacterial promoter is an example: although RNA polymerase covers >70 bp of DNA at initiation, the crucial sequences that it recognizes are the hexamers centered at V35 and V10.
A significant difference in gene organization between prokaryotes and eukaryotes is that structural genes in bacteria are organized in clusters, while those in eukaryotes occur individually. Clustering of structural genes allows them to be coordinately controlled by means of interactions at a single promoter: as a result of these interactions, the entire set of genes is either transcribed or not transcribed. In this chapter, we discuss this mode of control and its use by bacteria. The means employed to coordinate control of dispersed eukaryotic genes are discussed in 21 Regulation of transcription (for review see Miller and Reznikoff, 1978).
| Reviews | |
| Miller, J. and Reznikoff, W. (1978). The Operon. Cold Spring Harbor Symp. Quant. Biol.. | |
| Miller, J. and Reznikoff, W. (1978). The Operon. Cold Spring Harbor Symp. Quant. Biol.. | |
| Research | |
| Jacob, F. and Monod, J. (1961). Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318-389. | |