2. Structural gene clusters are coordinately controlled
10.2 Structural gene clusters are coordinately controlled |
| Key terms defined in this section |
| Operon is a unit of bacterial gene expression and regulation, including structural genes and control elements in DNA recognized by regulator gene product(s). |
Bacterial structural genes are often organized into clusters that include genes coding for proteins whose functions are related. It is common for the genes for the enzymes of a metabolic pathway to be organized into such a cluster. In addition to the enzymes actually involved in the pathway, other related activities may be included in the unit of coordinate control; for example, the protein responsible for transporting the small molecule substrate into the cell.
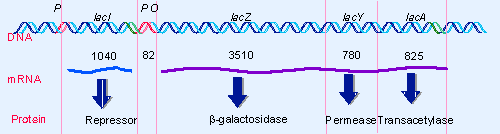 |
Figure 10.4 The lac operon occupies ~6000 bp of DNA. At the left the lacI gene has its own promoter and terminator. The end of the lacI region is adjacent to the promoter, P. The operator, O, occupies the first 26 bp of the long lacZ gene, followed by the lacY and lacA genes and a terminator. |
The cluster of the three lac structural genes, lacZYA, is typical. The protein products enable cells to take up and metabolize β-galactosides, such as lactose. Figure 10.4 summarizes the organization of the structural genes, their associated cis-acting regulatory elements, and the trans-acting regulatory gene. The roles of the three structural genes are:
- lacZ codes for the enzyme β-galactosidase, whose active form is a tetramer of ~500 kD. The enzyme breaks a β-galactoside into its component sugars. For example, lactose is cleaved into glucose and galactose (which are then further metabolized).
- lacY codes for the β-galactoside permease, a 30 kD membrane-bound protein constituent of the transport system. This transports β-galactosides into the cell.
- lacA codes for β-galactoside transacetylase, an enzyme that transfers an acetyl group from acetyl-CoA to β-galactosides.
Mutations in either lacZ or lacY can create the lac genotype, in which cells cannot utilize lactose. (The genotypic description "lac" without a qualifier indicates loss-of-function.) The lacZ mutations abolish enzyme activity, directly preventing metabolism of lactose. The lacY mutants cannot take up lactose from the medium. (No defect is identifiable in lacA cells, which is puzzling. It is possible that the acetylation reaction gives an advantage when the bacteria grow in the presence of certain analogs of β-galactosides that cannot be metabolized, because the modification results in detoxification and excretion.)
The entire system, including structural genes and the elements that control their expression, forms a common unit of regulation; this is called an operon. The activity of the operon is controlled by regulator gene(s), whose protein products interact with the cis-acting control elements.
We can distinguish between structural genes and regulator genes by the effects of mutations. A mutation in a structural gene deprives the cell of the particular protein for which the gene codes. But a mutation in a regulator gene influences the expression of all the structural genes that it controls. The consequences of a regulatory mutation reveal the type of regulation (Jacob and Monod, 1961; for review see Beckwith, 1978).
Transcription of the lacZYA genes is controlled by a regulator protein synthesized by the lacI gene. It happens that lacI is located adjacent to the structural genes, but it comprises an independent transcription unit with its own promoter and terminator. Since lacI specifies a diffusible product, in principle it need not be located near the structural genes; it can function equally well if moved elsewhere, or carried on a separate DNA molecule (the classic test for a trans-acting regulator).
The lac genes are controlled by negative regulation: they are transcribed unless turned off by the regulator protein. A mutation that inactivates the regulator causes the structural genes to remain in the expressed condition. The product of lacI is called the lac repressor, because its function is to prevent the expression of the structural genes.
The repressor is a tetramer of identical subunits of 38 kD each. There are ~10 tetramers in a wild-type cell. The regulator gene is transcribed into a monocistronic mRNA at a rate that appears to be governed simply by the affinity of its promoter for RNA polymerase.
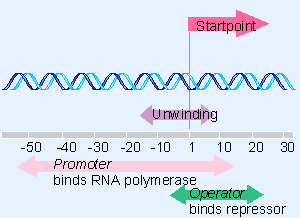 |
Figure 10.5 Repressor and RNA polymerase bind at sites that overlap around the startpoint of the lac operon. |
The repressor functions by binding to an operator (formally denoted Olac) at the start of the lacZYA cluster. The operator lies between the promoter (Plac) and the structural genes (lacZYA). When the repressor binds at the operator, it prevents RNA polymerase from initiating transcription at the promoter. Figure 10.5 expands our view of the region at the start of the lac structural genes. The operator extends from position -5 just upstream of the mRNA startpoint to position +21 within the transcription unit. So it overlaps the right end of the promoter. We discuss the relationship between repressor and RNA polymerase in more detail later (for review see Barkleya and Bourgeois, 1978; Beyreuther, 1978; Miller, 1978; Weber and Geisler, 1978).
| Reviews | |
| Barkleya, M. D. and Bourgeois, S. (1978). Repressor recognition of operator and effectors. In The Operon, Eds. Miller, J. and Reznikoff, W. Cold Spring Harbor Laboratory, New York 177-220. | |
| Beckwith, J. (1978). lac: the genetic system. In The Operon, Eds. Miller, J. and Reznikoff, W. Cold Spring Harbor Laboratory, New York 11-30. | |
| Beyreuther, K. (1978). Chemical structure and functional organization of lac repressor from E. coli. In The Operon, Eds. Miller, J. and Reznikoff, W. Cold Spring Harbor Laboratory, New York 123-154. | |
| Miller, J. H. (1978). The lacI gene: its role in lac operon control and its use as a genetic system. In The Operon, Eds. Miller, J. and Reznikoff, W. Cold Spring Harbor Laboratory, New York 31-88. | |
| Weber, K. and Geisler, N. (1978). Lac repressor fragments produced in vivo and in vitro: an approach to the understanding of the interaction of repressor and DNA. In The Operon, Eds. Miller, J. and Reznikoff, W. Cold Spring Harbor Laboratory, New York 155-176. | |
| Research | |
| Jacob, F. and Monod, J. (1961). Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318-389. | |