13. Attenuation can be controlled by translation
10.13 Attenuation can be controlled by translation |
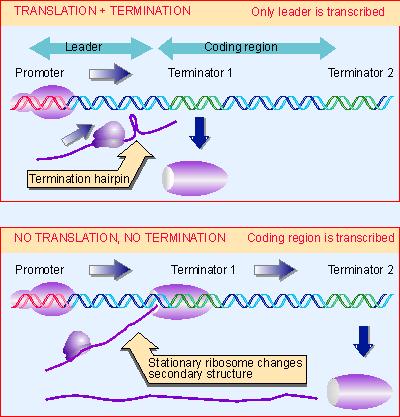 |
Figure 10.40 Termination can be controlled via changes in RNA secondary structure that are determined by ribosome movement. |
A more complex regulatory system is used in E. coli (where attenuation was originally discovered). The changes in secondary structure that control attenuation are determined by the position of the ribosome on mRNA. Figure 10.40 shows that termination requires that the ribosome can translate a leader segment of the mRNA. When the ribosome translates the leader region, a termination hairpin forms at terminator 1. But when the ribosome is prevented from translating the leader, the termination hairpin does not form, and RNA polymerase transcribes the coding region. This mechanism of antitermination therefore depends upon the ability of external circumstances to influence ribosome movement in the leader region (for review see Yanofsky, 1981).
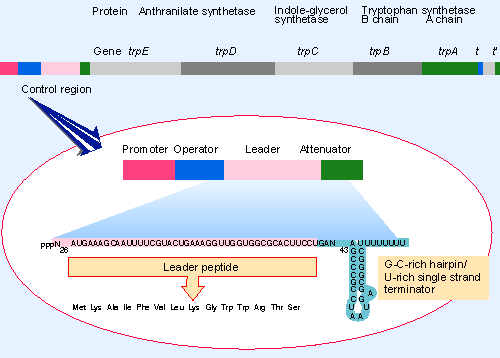 |
Figure 10.41 The trp operon consists of five contiguous structural genes preceded by a control region that includes a promoter, operator, leader peptide coding region, and attenuator. |
The trp operon consists of five structural genes arranged in a contiguous series, coding for the three enzymes that convert chorismic acid to tryptophan. Figure 10.41 shows that transcription starts at a promoter at the left end of the cluster. trp operon expression is controlled by two separate mechanisms. Repression of expression is exercised by a repressor protein (coded by the unlinked gene trpR) that binds to an operator that is adjacent to the promoter. Attenuation controls the progress of RNA polymerase into the operon by regulating whether termination occurs at a site preceding the first structural gene.
Attenuation was first revealed by the observation that deleting a sequence between the operator and the trpE coding region can increase the expression of the structural genes. This effect is independent of repression: both the basal and derepressed levels of transcription are increased. So this site influences events that occur after RNA polymerase has set out from the promoter (irrespective of the conditions prevailing at initiation).
The sequence between the promoter and the trpE gene has two interesting features:
- A short coding sequence could represent a leader peptide of 14 amino acids.
- The attenuator (intrinsic terminator) provides a barrier to transcription into the structural genes. RNA polymerase terminates there, either in vivo or in vitro, to produce a 140-base transcript.
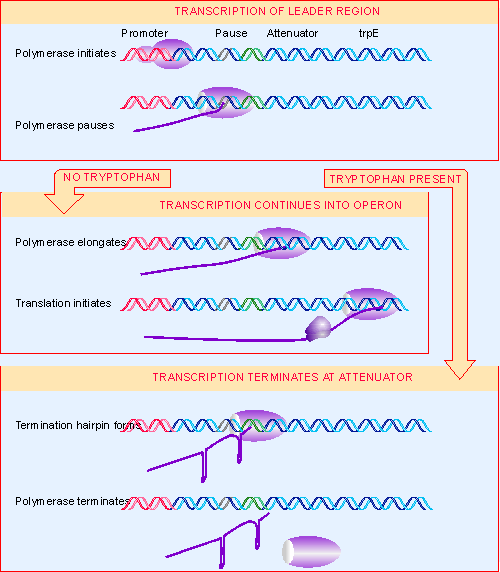 |
Figure 10.42 An attenuator controls the progression of RNA polymerase into the trp genes. RNA polymerase initiates at the promoter and then proceeds to position 90, where it pauses before proceeding to the attenuator at position 140. In the absence of tryptophan, the polymerase continues into the structural genes (trpE starts at +163). In the presence of tryptophan there is ~90% probability of termination to release the 140-base leader RNA. |
Termination at the attenuator responds to the level of tryptophan, as illustrated in Figure 10.42. In the presence of adequate amounts of tryptophan, termination is efficient. But in the absence of tryptophan, RNA polymerase can continue into the structural genes.
Repression and attenuation respond in the same way to the level of tryptophan. When tryptophan is present, the operon is repressed; and most of the RNA polymerases that escape from the promoter then terminate at the attenuator. When tryptophan is removed, RNA polymerase has free access to the promoter, and also is no longer compelled to terminate prematurely.
Attenuation has ~10 effect on transcription. When tryptophan is present, termination is effective, and the attenuator allows only ~10% of the RNA polymerases to proceed. In the absence of tryptophan, attenuation allows virtually all of the polymerases to proceed. Together with the ~70 increase in initiation of transcription that results from the release of repression, this allows an ~700-fold range of regulation of the operon.
How can termination of transcription at the attenuator respond to the level of tryptophan? The sequence of the leader region suggests a mechanism. Figure 10.41 shows that it contains a ribosome binding site whose AUG codon is followed by a short coding region that contains two successive codons for tryptophan. When the cell runs out of tryptophan, ribosomes initiate translation of the leader peptide, but stop when they reach the Trp codons. The sequence of the mRNA suggests that this ribosome stalling influences termination at the attenuator.
The leader sequence can be written in alternative base-paired structures. The ability of the ribosome to proceed through the leader region controls transitions between these structures. The structure determines whether the mRNA can provide the features needed for termination.
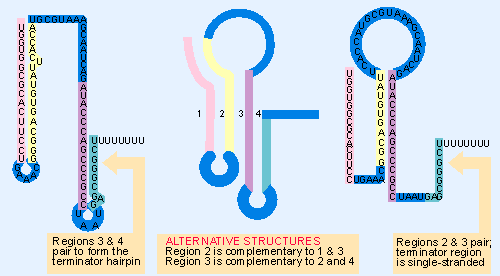 |
Figure 10.43 The trp leader region can exist in alternative base-paired conformations. The center shows the four regions that can base pair. Region 1 is complementary to region 2, which is complementary to region 3, which is complementary to region 4. On the left is the conformation produced when region 1 pairs with region 2, and region 3 pairs with region 4. On the right is the conformation when region 2 pairs with region 3, leaving regions 1 and 4 unpaired. |
Figure 10.43 draws these structures. In the first, region 1 pairs with region 2; and region 3 pairs with region 4. The pairing of regions 3 and 4 generates the hairpin that precedes the U8 sequence: this is the essential signal for intrinsic termination. Probably the RNA would take up this structure in lieu of any outside intervention.
A different structure is formed if region 1 is prevented from pairing with region 2. In this case, region 2 is free to pair with region 3. Then region 4 has no available pairing partner; so it is compelled to remain single-stranded. So the terminator hairpin cannot be formed.
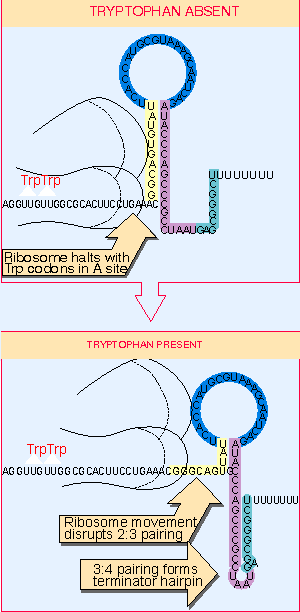 |
Figure 10.44 The alternatives for RNA polymerase at the attenuator depend on the location of the ribosome, which determines whether regions 3 and 4 can pair to form the terminator hairpin. |
Figure 10.44 shows that the position of the ribosome can determine which structure is formed, in such a way that termination is attenuated only in the absence of tryptophan. The crucial feature is the position of the Trp codons in the leader peptide coding sequence.
When tryptophan is present, ribosomes are able to synthesize the leader peptide. They continue along the leader section of the mRNA to the UGA codon, which lies between regions 1 and 2. As shown in the lower part of the figure, by progressing to this point, the ribosomes extend over region 2 and prevent it from base pairing. The result is that region 3 is available to base pair with region 4, generating the terminator hairpin. Under these conditions, therefore, RNA polymerase terminates at the attenuator.
When there is no tryptophan, ribosomes stall at the Trp codons, which are part of region 1, as shown in the upper part of the figure. So region 1 is sequestered within the ribosome and cannot base pair with region 2. This means that regions 2 and 3 become base paired before region 4 has been transcribed. This compels region 4 to remain in a single-stranded form. In the absence of the terminator hairpin, RNA polymerase continues transcription past the attenuator (Lee and Yanofsky, 1977; Zurawski et al., 1978; for review see Landick and Yanofsky, 1987).
Control by attenuation requires a precise timing of events. For ribosome movement to determine formation of alternative secondary structures that control termination, translation of the leader must occur at the same time when RNA polymerase approaches the terminator site. A critical event in controlling the timing is the presence of a site that causes the RNA polymerase to pause at base 90 along the leader. The RNA polymerase remains paused until a ribosome translates the leader peptide. Then the polymerase is released and moves off toward the attenuation site. By the time it arrives there, secondary structure of the attenuation region has been determined.
By providing a mechanism to sense the inadequacy of the supply of Trp-tRNA, attenuation responds directly to the need of the cell for tryptophan in protein synthesis (for review see Yanofsky and Crawford, 1987).
How widespread is the use of attenuation as a control mechanism for bacterial operons? It is used in at least six operons that code for enzymes concerned with the biosynthesis of amino acids. So a feedback from the level of the amino acid available for protein synthesis (as represented by the availability of aminoacyl-tRNA) to the production of the enzymes may be common (for review see Bauer et al., 1983).
The use of the ribosome to control RNA secondary structure in response to the availability of an aminoacyl-tRNA establishes an inverse relationship between the presence of aminoacyl-tRNA and the transcription of the operon, equivalent to a situation in which aminoacyl-tRNA functions as a corepressor of transcription. Since the regulatory mechanism is mediated by changes in the formation of duplex regions, attenuation provides a striking example of the importance of secondary structure in the termination event, and of its use in regulation.
This section updated 1-7-2000
| Reviews | |
| Bauer, C. E. et al. (1983). Attenuation in bacterial operons. In Gene Function in Prokaryotes, Eds. J. Beckwith, J. E. Davies, and J. A. Gallant. Cold Spring Harb 65-89. | |
| Landick, R. and Yanofsky, C. (1987). Transcription attenuation In E. coli and S. typhimurium. In E. coli and S. typhimurium, Ed. F. C. Neidhardt, American Society for Microbiology, Washington DC 1276-1301. | |
| Yanofsky, C. (1981). Attenuation in the control of expression of bacterial operons. Nature 289, 751-758. | |
| Yanofsky, C. and Crawford, I. P. (1987). The tryptophan operon. In E. coli and S. typhimurium, Ed. F. C. Neidhardt, American Society for Microbiology, Washington DC 1453-1472. | |
| Research | |
| Lee, F. and Yanofsky, C. (1977). Transcription termination at the trp operon attenuators of E. coli and S. typhimurium: RNA secondary structure and regulation of termination. Proc. Nat. Acad. Sci. USA 74, 4365-4368. | |
| Zurawski, G. et al. (1978). Translational control of transcription termination at the attenuator of the E. coli tryptophan operon. Proc. Nat. Acad. Sci. USA 75, 5988-5991. | |