5. Promoter recognition depends on consensus sequences
9.5 Promoter recognition depends on consensus sequences |
| Key terms defined in this section |
| Consensus sequence is an idealized sequence in which each position represents the base most often found when many actual sequences are compared. |
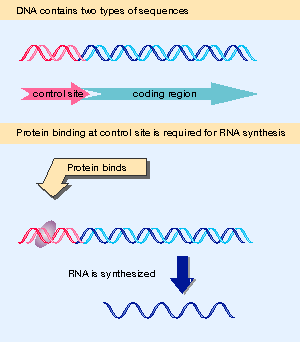 |
Figure 1.33 Control sites in DNA provide binding sites for proteins; coding regions are expressed via the synthesis of RNA. |
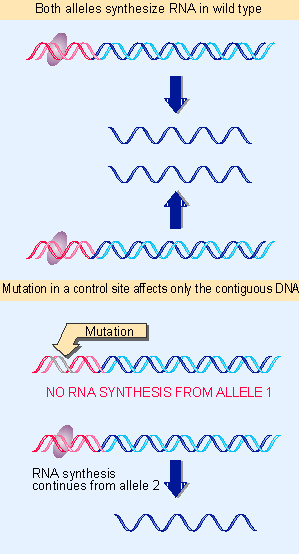 |
Figure 1.34 A cis-acting site controls the adjacent DNA but does not influence the other allele. |
As a sequence of DNA whose function is to be recognized by proteins, a promoter differs from sequences whose role is to be transcribed or translated. The information for promoter function is provided directly by the DNA sequence: its structure is the signal. This is a classic example of a cis-acting site, as defined previously in Figure 1.33 and Figure 1.34. By contrast, expressed regions gain their meaning only after the information is transferred into the form of some other nucleic acid or protein.
A key question in examining the interaction between an RNA polymerase and its promoter is how the protein recognizes a specific promoter sequence. Does the enzyme have an active site that distinguishes the chemical structure of a particular sequence of bases in the DNA double helix? How specific are its requirements?
One way to design a promoter would be for a particular sequence of DNA to be recognized by RNA polymerase. Every promoter would consist of, or at least include, this sequence. In the bacterial genome, the minimum length that could provide an adequate signal is 12 bp. (Any shorter sequence is likely to occur Xjust by chance Xa sufficient number of additional times to provide false signals. The minimum length required for unique recognition increases with the size of genome.) The 12 bp sequence need not be contiguous. If a specific number of base pairs separates two constant shorter sequences, their combined length could be less than 12 bp, since the distance of separation itself provides a part of the signal (even if the intermediate sequence is itself irrelevant).
Attempts to identify the features in DNA that are necessary for RNA polymerase binding started by comparing the sequences of different promoters. Any essential nucleotide sequence should be present in all the promoters. Such a sequence is said to be conserved. However, a conserved sequence need not necessarily be conserved at every single position; some variation is permitted. How do we analyze a sequence of DNA to determine whether it is sufficiently conserved to constitute a recognizable signal?
Putative DNA recognition sites can be defined in terms of an idealized sequence that represents the base most often present at each position. A consensus sequence is defined by aligning all known examples so as to maximize their homology. For a sequence to be accepted as a consensus, each particular base must be reasonably predominant at its position, and most of the actual examples must be related to the consensus by rather few substitutions, say, no more than 1 V2.
The striking feature in the sequence of promoters in E. coli is the lack of any extensive conservation of sequence over the 60 bp associated with RNA polymerase. The sequence of much of the binding site is irrelevant. But some short stretches within the promoter are conserved, and they are critical for its function. Conservation of only very short consensus sequences is a typical feature of regulatory sites (such as promoters) in both prokaryotic and eukaryotic genomes.
There are four conserved features in a bacterial promoter: the startpoint; the V10 sequence; the V35 sequence; and the separation between the V10 and V35 sequences:
- The startpoint is usually (>90% of the time) a purine. It is common for the startpoint to be the central base in the sequence CAT, but the conservation of this triplet is not great enough to regard it as an obligatory signal.
- Just upstream of the startpoint, a 6 bp region is recognizable in almost all promoters. The center of the hexamer generally is close to 10 bp upstream of the startpoint; the distance varies in known promoters from position -18 to -9. Named for its location, the hexamer is often called the V10 sequence. Its consensus is TATAAT, and can be summarized in the form
T80 A95 T45 A60 A50 T96
where the subscript denotes the percent occurrence of the most frequently found base, varying from 45-96%. (A position at which there is no discernible preference for any base would be indicated by N.) If the frequency of occurrence indicates likely importance in binding RNA polymerase, we would expect the initial highly conserved TA and the final almost completely conserved T in the V10 sequence to be the most important bases.
- Another conserved hexamer is centered ~35 bp upstream of the startpoint. This is called the V35 sequence. The consensus is TTGACA; in more detailed form, the conservation is
T82 T84 G78 A65 C54 A45
- The distance separating the V35 and V10 sites is between 16-18 bp in 90% of promoters; in the exceptions, it is as little as 15 or as great as 20 bp. Although the actual sequence in the intervening region is unimportant, the distance is critical in holding the two sites at the appropriate separation for the geometry of RNA polymerase.
 |
Figure 9.14 A typical promoter has three components, consisting of consensus sequences at -35 and -10, and the startpoint. |
The optimal promoter is a sequence consisting of the V35 hexamer, separated by 17 bp from the V10 hexamer, lying 7 bp upstream of the startpoint. The structure of a promoter, showing the permitted range of variation from this optimum, is illustrated in Figure 9.14.
A major source of information about promoter function is provided by mutations. Mutations in promoters affect the level of expression of the gene(s) they control, without altering the gene products themselves. Most are identified as bacterial mutants that have lost, or have very much reduced, transcription of the adjacent genes. They are known as down mutations. Less often, mutants are found in which there is increased transcription from the promoter. They have up mutations.
It is important to remember that "up" and "down" mutations are defined relative to the usual efficiency with which a particular promoter functions. This varies widely. So a change that is recognized as a down mutation in one promoter might never have been isolated in another (which in its wild-type state could be even less efficient than the mutant form of the first promoter). Information gained from studies in vivo simply identifies the overall direction of the change caused by mutation.
Is the most effective promoter one that has the actual consensus sequences? This expectation is borne out by the simple rule that up mutations usually increase homology with one of the consensus sequences or bring the distance between them closer to 17 bp. Down mutations usually decrease the resemblance of either site with the consensus or make the distance between them more distant from 17 bp. Down mutations tend to be concentrated in the most highly conserved positions, which confirms their particular importance as the main determinant of promoter efficiency. However, occasional exceptions to these rules demonstrate that promoter efficiency cannot be predicted entirely from conformity to the consensus.
To determine the absolute effects of promoter mutations, we must measure the affinity of RNA polymerase for wild-type and mutant promoters in vitro. There is ~100-fold variation in the rate at which RNA polymerase binds to different promoters in vitro, which correlates well with the frequencies of transcription when their genes are expressed in vivo. Taking this analysis further, we can investigate the stage at which a mutation influences the capacity of the promoter. Does it change the affinity of the promoter for binding RNA polymerase? Does it leave the enzyme able to bind but unable to initiate? Is the influence of an ancillary factor altered?
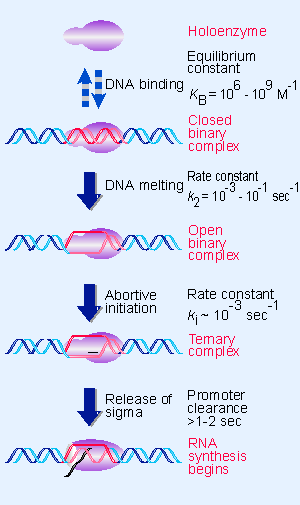 |
Figure 9.9 RNA polymerase passes through several steps prior to elongation. A closed binary complex is converted to an open form and then into a ternary complex. |
By measuring the kinetic constants for formation of a closed complex and its conversion to an open complex, as defined in Figure 9.9, we can dissect the two stages of the initiation reaction:
- Down mutations in the V35 sequence reduce the rate of closed complex formation (they reduce KB), but do not inhibit the conversion to an open complex.
- Down mutations in the V10 sequence do not affect the initial formation of a closed complex, but they slow its conversion to the open form (they reduce k2).
These results suggest that the function of the V35 sequence is to provide the signal for recognition by RNA polymerase, while the V10 sequence allows the complex to convert from closed to open form. We might view the V35 sequence as comprising a "recognition domain," while the V10 sequence comprises an "unwinding domain" of the promoter.
The consensus sequence of the V10 site consists exclusively of A PT base pairs, which assists the initial melting of DNA into single strands. The lower energy needed to disrupt A PT pairs compared with G PC pairs means that a stretch of A PT pairs demands the minimum amount of energy for strand separation.
The sequence immediately around the startpoint influences the initiation event. And the initial transcribed region (from +1 to +30) influences the rate at which RNA polymerase clears the promoter, and therefore has an effect upon promoter strength. So the overall strength of a promoter cannot be predicted entirely from its V35 and V10 consensus sequences.
A "typical" promoter relies upon its V35 and V10 sequences to be recognized by RNA polymerase, but one or the other of these sequences can be absent from some (exceptional) promoters. In at least some of these cases, the promoter cannot be recognized by RNA polymerase alone, and the reaction requires the intercession of ancillary proteins, which overcome the deficiency in intrinsic interaction between RNA polymerase and the promoter.