2.3 Examples of Molecular-Recognition Processes
|
2.3 Examples of Molecular-Recognition Processes
Most naturally occurring systems that demonstrate recognition (an enzyme recognizing its substrate molecule) involve many binding features and are too complicated to adequately model. Hence, in order to explore the basis of recognition, researchers in the field have first focused their efforts on much simpler systems for actual design, with the goal of investigating cases of increasing complexity. Much molecular recognition work has been done with macropolycyclic structures—molecules containing many branches of linked atoms—because of their suitability when designing artificial receptors. They are large (macro) and may therefore contain cavities of appropriate size and shape to contain a cation, anion, or more complex molecules.[2] Macropolycyclic structures possess numerous branches, bridges, and connections (polycyclic) that allow them to maintain the shape and to confer specific dynamic features. Figure 2.1 shows examples of macrocyclic and higher order structures.
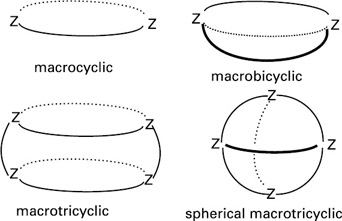
Figure 2.1: Some macropolycyclic structures. (From Lehn, 1973. With permission from Springer-Verlag 1973.)
Also, such molecules provide means for arranging geometrically the desired structural groups, binding sites, and reactive functions. When a substrate is bound into the cavity, the result is an inclusion complex, a cryptate designated by the mathematical inclusion sign ⊂: (substrate ⊂ receptor).
Exoreceptors: Outwardly Directed Molecular Recognition
Most biological examples of molecular recognition are analogous to enzymes, where the active sites are contained inside the cavity of a large protein molecule, which then binds a smaller substrate fitting into the cleft (endorecognition). Most of the work with macrocyclic/macropolycyclic architectures has been along these lines.
The opposite procedure consists of making use of an external surface with protuberances and depressions. The receptor then binds to the substrate by surface-tosurface interaction—analogous to what happens in protein-protein interactions and considered to be an outwardly directed recognition process (exorecognition). Strong and selective binding requires a large enough contact area and a sufficient number of interfaces, as well as geometrical and site complementarity, as mentioned above. Another example of exoreceptor recognition involves a molecule (ligand) containing recognition sites wrapping itself around a central metal ion. A concrete example of this will be given in the last section.
Most molecular recognition at surfaces and interfaces (monolayers, films, membranes, cell walls, organic or inorganic solids) involves outside-directed recognition sites.
Examples of Simple Molecular Recognition: Spheres, Lines, and Tetrahedra
The simplest recognition process is that of spherical substrates, because the sphere is the simplest object in three-dimensional space. These are either positive-charged metal cations (alkali, alkaline-earth, and lanthanide cations) or the negative halide anions. The receptors that recognize such species fall into three main classes: (1) natural macrocycles such as valinomycin; (2) synthetic macrocyclic polyethers, the crown ethers (and derivatives); and (3) synthetic macropolycyclic ligands, the cryptands and other types such as the cryptospherands. The major difference between the macrocycles and macropolycycles is that whereas the former have their main feature as that of a ring into which the cation is bound, macropolycycles possess by nature a three-dimensional cavity, thus allowing, in principle, a much better complementary inclusion of the substrate. Binding into a ring cavity as is found in a macrocycle does not necessarily mean binding to the "equator" of the cation only. The flexibility of the ring can allow adaptation of the ligand—as in the cubic binding of K+ into valinomycin (figure 2.2) (Dobler 1981). As an example of spherical recognition, macrobicyclic ligands, as shown in figure 2.3, form cryptates by inclusion of a metal cation inside the molecule. This class of molecular structures shows pronounced selectivity as a function of the size complementarity between the cation and the intramolecular cavity—a feature termed spherical recognition. As the bridges of the surrounding molecule are lengthened, the size of the cavity increases gradually, with the most strongly bound ion becoming respectively Li+, Na+, and then K+ (Lehn and Sauvage 1975; Lehn 1978). Thus these ligands present peak selectivity, being able to discriminate against cations that are either smaller or larger than their cavity. For more flexible cryptands containing longer chains, the cavities are larger and more adjustable. Thermodynamics and energetics make, of course, the question of binding more complicated than simply one of geometrical fitting. (For a review of thermodynamics data on macrocycle interactions with cations, anions, and neutral molecules, see Izatt et al. 1995.)
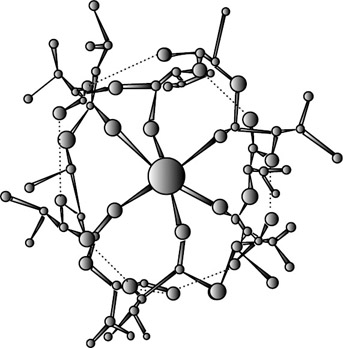
Figure 2.2: Valinomycin with K+. (From Ionophores and Their Structures, M. Dobler. 1981, John Wiley. Reprinted by permission of John Wiley & Sons, Inc.)
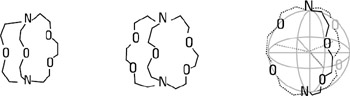
Figure 2.3: Three different examples of macrobicyclic ligands. As the size of the internal cavity gradually increases from left to right, the most strongly bound ion changes from Li+ to Na+ to K+.
Tetrahedral Recognition
Selective binding of tetrahedral substrates requires the construction of a receptor molecule with a tetrahedral recognition site. This may be achieved by positioning four suitable binding sites at the corners of a tetrahedron and maintaining them in the appropriate position with six bridges. Such a structure has been realized in a spherical macrotricyclic cryptand (Graf and Lehn 1975), which contains four nitrogens located at the corners of a tetrahedron and six oxygens located at the corners of an octahedron. This binds a tetrahedral NH4+ cation exceptionally strongly and selectively (relative to K+, for example), forming the ammonium cryptate as shown in figure 2.4 (Graf et al. 1982; Dietrich et al. 1987). This complex presents a high degree of structural (shape and size) and interaction site complementarity between the substrate NH4+ and the receptor. The ammonium ion fits exactly into the cavity and is held by a tetrahedral array of +N-H…N hydrogen bonds and by electrostatic interactions with the six oxygens. It should be pointed out that the molecule is firmly held inside the cavity and does not undergo internal rotation—as happens in a similar macrobicyclic molecule (figure 2.5), with which NH4+ can also form a cryptate (Dietrich et al. 1987).
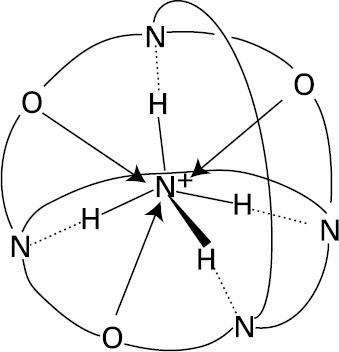
Figure 2.4: Ammonium cryptate. (Reprinted with permission from Graf et al. 1982. 1982 American Chemical Society.)
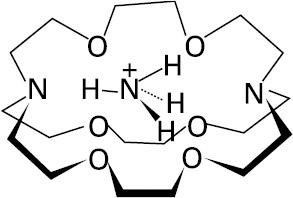
Figure 2.5: Ammonia in macrobicyclic receptor molecule. (Reprinted with permission from Dietrich et al. 1987. 1987 American Chemical Society.)
Other Types of Molecules That Can Be Bound
The binding and recognition of neutral molecules make use of electrostatic, donoracceptor, and especially hydrogen bonding interactions. Of special interest is the use of hydrogen bonding between polar sites to bind to the substrate because of the ubiquitousness of hydrogen bonding in organic systems. Here, substrate recognition results from the formation of specific hydrogen bonding patterns between complementary subunits, in a way reminiscent of base pairing in nucleic acids. Such groups have been positioned in acyclic or macrocyclic receptors, respectively defining clefts or cavities into which binding of substrates of complementary structures has been shown to take place. Figure 2.6 (Rebek 1988 and following references) illustrates the formation of a complex through hydrogen bonding of adenine in a cleft. Hydrogen bonding plays a major role in the recognition of nucleic acid sequences by specially designed synthetic molecules or by proteins, as well as in the recognition of oligosaccharides by proteins.
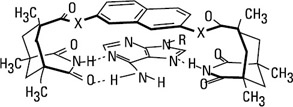
Figure 2.6: Binding of adenine in a cleft. (From Rebek, 1990. With permission from Accounts Chem. Res., 1987.)
Anion Recognition
Molecular recognition investigations focused first on the complexation of metal ions and of cationic molecules. But anionic species play a very important role in chemistry and in biology. Research has become increasingly active in the area of anion complexation and recognition, involving the development of anion receptor molecules and binding subunits for anionic functional groups. Anionic substrates are large compared to cations, and possess a wide range of geometries (spherical, linear, planar, tetrahedral, and octahedral). Much work has been done on the binding of carboxylates and phosphates. The design of receptor units for these functional groups is of special interest because they serve as anchoring sites for numerous biological substrates. The strong complexation of adenosine mono-, di-, and triphosphates (AMP, ADP, and ATP) is particularly significant in view of their role in bioenergetics. Other areas of research are the development of cyclic analogues of biological polyamines (which can interact with biomolecules) and work on creating receptors containing the guanidinium group. Much attention has been paid to developing chiral receptors—that is, the molecule can come in two forms, a "lefthand" and a "right-hand" form. Such chiral receptors allow for chiral discrimination of the target molecules, analogous to what happens in biological systems.
Linear Recognition
Linear recognition can be performed with a receptor presenting two recognition sites at the opposite ends of a cavity, with the length of the cavity providing a filter as to the length of the substrate molecule that will be preferentially accepted. An example is an ellipsoidal cryptand bis-tren, as shown in figure 2.7, which strongly and selectively binds N−3 due to its size, shape, and site complementarity (Dietrich et al. 1984; Lehn, Sonveaux, and Willard 1978). This cryptand, although it also binds fluoride and chloride, does so much less well and with appreciable distortions of the ligand; it much prefers a linear molecule of a size compatible with the size of the cavity.
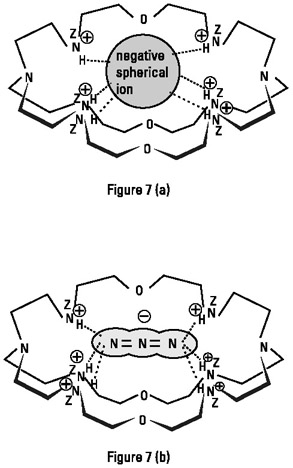
Figure 2.7: (a) Hexaprotonated form of an ellipsoidal polyammonium bis-tren macrobicycle, in this case binding a spherical halide ion. (from Dietrich et al. 1984. With permission from Helv. Chim. Acta, 1984.) (b) Hexaprotonated form of an ellipsoidal polyammonium bis-tren macrobicycle, binding the linear triatomic anion N3−. (Reprinted with permission from Lehn, Sonveaux, and Willard 1978. 1978 American Chemical Society.)
Another example of what could be termed linear recognition is in the hydrolysis of N-acetylglucosamine(NAG) by lysozyme. The hexamer (NAG)6 is the shortest NAG oligosaccharide that is hydrolyzed rapidly. It turns out that there exist six sites on the catalyst that must be filled very specifically, each site recognizing a different part of the hexamer monomer. Anything shorter will not be sufficient (Gates 1992: 162).
[2]To give an idea on size, most cavities range from approximately 2 to approximately 15 angstroms.
|