10. Three codons terminate protein synthesis
6.10 Three codons terminate protein synthesis |
| Key terms defined in this section |
| Missense mutations change a single codon and so may cause the replacement of one amino acid by another in a protein sequence. Nonsense codon means a termination codon. Termination codon is one of three (UAG, UAA, UGA) that causes protein synthesis to terminate. |
Only 61 triplets are assigned to amino acids. The other three triplets are termination codons (or stop codons) that end protein synthesis. They have casual names from the history of their discovery. The UAG triplet is called the amber codon; UAA is the ochre codon; and UGA is sometimes called the opal codon.
The nature of these triplets was originally shown by a genetic test that distinguished two types of point mutation:
- A point mutation that changes a codon to represent a different amino acid is called a missense mutation. One amino acid replaces the other in the protein; the effect on protein function depends on the site of mutation and the nature of the amino acid replacement.
- When a point mutation creates one of the three termination codons, it causes premature termination of protein synthesis at the mutant codon. This is likely to abolish protein function, since only the first part of the protein is made in the mutant cell. A change of this sort is called a nonsense mutation.
(Sometimes the term nonsense codon is used to describe the termination triplets. "Nonsense" is really a misnomer, since the codons do have meaning, albeit a disruptive one in a mutant gene.)
In every gene that has been sequenced, one of the termination codons lies immediately after the codon representing the C-terminal amino acid of the wild-type sequence. Nonsense mutations show that any one of the three codons is sufficient to terminate protein synthesis within a gene. The UAG, UAA, and UGA triplet sequences are therefore necessary and sufficient to end protein synthesis, whether occurring naturally at the end of a gene or created by mutation within a coding sequence.
In bacterial genes, UAA is the most commonly used termination codon. UGA is used more heavily than UAG, although there appear to be more errors reading UGA. (An error in reading a termination codon, when an aminoacyl-tRNA improperly responds to it, results in the continuation of protein synthesis until another termination codon is encountered.)
Two stages are involved in ending translation. The termination reaction itself involves release of the protein chain from the last tRNA. The post-termination reaction involves release of the tRNA and mRNA, and dissociation of the ribosome into its subunits.
None of the termination codons is represented by a tRNA. They function in an entirely different manner from other codons, and are recognized directly by protein factors. (Since the reaction does not depend on codon-anticodon recognition, there seems to be no particular reason why it should require a triplet sequence. Presumably this reflects the evolution of the genetic code.)
In E. coli two related proteins catalyze termination. They are called release factors (RF), and are specific for different sequences (927). RF-1 recognizes UAA and UAG; RF-2 recognizes UGA and UAA. The factors act at the ribosomal A site and require polypeptidyl-tRNA in the P site. The release factors are present at much lower levels than initiation or elongation factors; there are ~600 molecules of each per cell, equivalent to 1 RF per 10 ribosomes. Probably at one time there was only a single release factor, recognizing all termination codons, and later it evolved into two factors with specificities for particular codons.
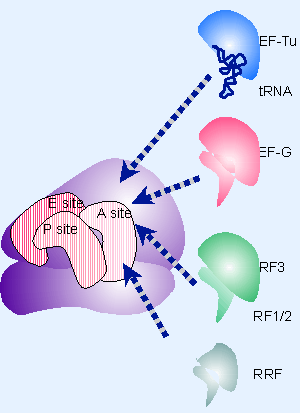 |
Figure 6.27 Molecular mimicry enables the elongation factor Tu-tRNA complex, the translocation factor EF-G, and the release factors RF1/2-RF3 to bind to the same ribosomal site. |
RF1 and RF2 recognize the termination codons and activate the ribosome to hydrolyze the peptidyl tRNA. Cleavage of polypeptide from tRNA takes place by a reaction analogous to the usual peptidyl transfer, except that the acceptor is H2O instead of aminoacyl-tRNA. Then RF1 or RF2 is released from the ribosome by RF-3, which is a GTP-binding protein related to EF-G (1161, 1162). RF3 resembles the GTP-binding domains of EF-Tu and EF-G, and RF1/2 resemble the C-terminal of EF-G, which mimics tRNA. This suggests that the action of RF3 on RF1/2 utilize the same site that is used by the elongation factors. Figure 6.27 illustrates the basic idea that these factors all have the same general shape and bind to the ribosome successively at the same site (basically the A site or a region extensively overlapping with it).
Mutations in the RF genes reduce the efficiency of termination, as seen by an increased ability to continue protein synthesis past the termination codon. Over-expression of RF-1 or RF-2 increases the efficiency of termination at the codons on which it acts. This suggests that codon recognition by RF1 or RF2 competes with aminoacyl-tRNAs that erroneously recognize the termination codons. The release factors recognize their target sequences very efficiently (1163).
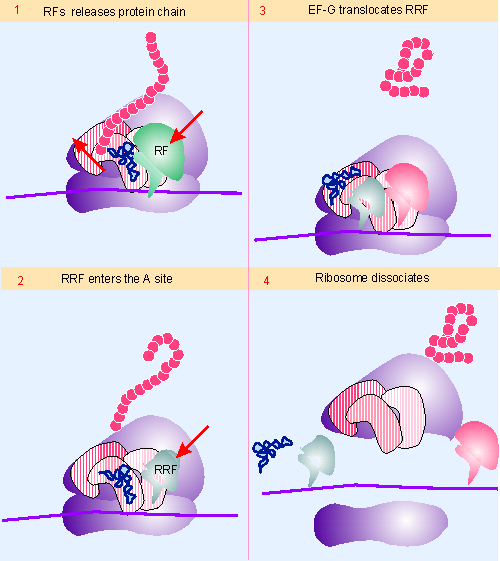 |
Figure 6.28 The RF (release factor) terminates protein synthesis by releasing the protein chain. The RRF (ribosome recycling factor) releases the last tRNA, and EF-G releases RRF, causing the ribosome to dissocuate. Animated figure |
The termination reaction involves release of the completed polypeptide, but leaves a deacylated tRNA and the mRNA still associated with the ribosome. Figure 6.28 shows that the dissocation of the remaining components (tRNA, mRNA, 30S and 50S subunits) requires the factor RRF, ribosome recycling factor. This acts together with EF-G in a reaction that uses hydrolysis of GTP. Like the other factors involved in release, RRF has a structure that mimics tRNA, except that it lacks an equivalent for the 3′ amino acid-binding region (926). IF-3 is also required, which brings the wheel full circle to its original discovery, when it was proposed to be a dissociation factor! RRF acts on the 50S subunit, and IF-3 acts to remove deacaylated tRNA from the 30S subunit. Once the subunits have separated, IF-3 remains necessary, of course, to prevent their reassociation.
This section updated 1-10-2000
| Research | |
| 926: | Selmer, M. et al. (1999). Crystal structure of Thermotogo maritima ribosome recycling factor: a tRNA mimic.. Science 286, 2349-2352. |
| 927: | Scolnick, E. et al. (1968). Release factors differing in specificity for terminator codons.. Proc. Nat. Acad. Sci. USA 61, 768-774. |
| 1161: | Milman, G. , Goldstein, J. , Scolnick, E. , and Caskey, T. (1969). Peptide chain termination. 3. Stimulation of in vitro termination.. Proc. Nat. Acad. Sci. USA 63, 183-190. |
| 1162: | Mikuni, O. , Ito, K. , Moffat, J. , Matsumura, K. , McCaughan, K. , Nobukuni, T. , Tate, W. , and Nakamura, Y. (1994). Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli.. Proc. Nat. Acad. Sci. USA 91, 5798-5802. |
| 1163: | Freistroffer, D. V. , Kwiatkowski, M. , Buckingham, R. H. , and Ehrenberg, M. (2000). The accuracy of codon recognition by polypeptide release factors.. Proc. Nat. Acad. Sci. USA 97, 2046-2051. |