10. Histone acetylation and deacetylation control chromatin activity
21.10 Histone acetylation and deacetylation control chromatin activity |
| Key terms defined in this section |
| HAT (histone acetyltransferase) enzymes modify histones by addition of acetyl groups; some transcriptional coactivators have HAT activity. HDAC (histone deacetyltransferase) enzymes remove acetyl groups from histones; they may be associated with repressors of transcription. |
It has long been known that the state of histone acetylation is correlated with the state of gene expression. All the core histones are acetylated. Histone acetylation appears to be increased in a domain containing active genes, and acetylated chromatin is more sensitive to DNAase I and (possibly) to micrococcal nuclease. Histone acetylation also occurs during S phase, when it may be associated with the incorporation of histones into nucleosomes. The most striking change in histone modification found to date is the underacetylation of histone H4 on the inactive X chromosome in female mammals. This suggests that absence of acetyl groups may be a prerequisite for a more condensed, inactive structure.
Enzymes that can acetylate histones are called histone acetyltransferases or HATs; the acetyl groups are removed by histone deacetylases or HDACs. There are two groups of HAT enzymes: group A describes those that are involved with transcription; group B describes those involved with nucleosome assembly. Two inhibitors have been useful in analyzing acetylation. Trichostatin and butyric acid inhibit histone deacetylases, and cause acetylated nucleosomes to accumulate. The use of these inhibitors has supported the general view that acetylation is associated with gene expression; in fact, the ability of butyric acid to cause changes in chromatin resembling those found upon gene activation was one of the first indications of the connection between acetylation and gene activity.
The breakthrough in analyzing the role of histone acetylation was provided by the characterization of the acetylating and deacetylating enzymes, and their association with other proteins that are involved in specific events of activation and repression. A basic change in our view of histone acetylation was caused by the discovery that HATs are not necessarily dedicated enzymes associated with chromatin: rather it turns out that known activators of transcription have HAT activity.
The connection was established when the catalytic subunit of a group A HAT was identified as a homologue of the yeast regulator protein GCN5. Then it was shown that GCN5 itself has HAT activity (with histones H3 and H4 as substrates). GCN5 is part of an adaptor complex that is necessary for the interaction between certain enhancers and their target promoters. Its HAT activity is required for activation of the target gene (Brownell et al., 1996).
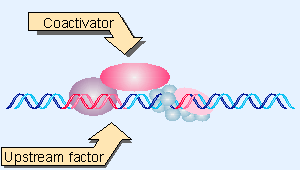 |
Figure 20.26 An upstream transcription factor may bind a coactivator that contacts the basal apparatus. |
One of the first general activators to be characterized as an HAT was p300/CBP. (Actually, p300 and CBP are different proteins, but they are so closely related that they are often referred to as a single type of activity.) p300/CBP is a coactivator that links an upstream transcription factor to the basal apparatus (see Figure 20.26). p300/CBP interacts with various transcription factors, including hormone receptors, AP-1 (c-Jun and c-Fos), and MyoD. The interaction is inhibited by the viral regulator proteins adenovirus E1A and SV40 T antigen, which bind to p300/CBP to prevent the interaction with transcription factors; this explains how these viral proteins inhibit cellular transcription. (This inhibition is important for the ability of the viral proteins to contribute to the tumorigenic state; see 28 Oncogenes and cancer).
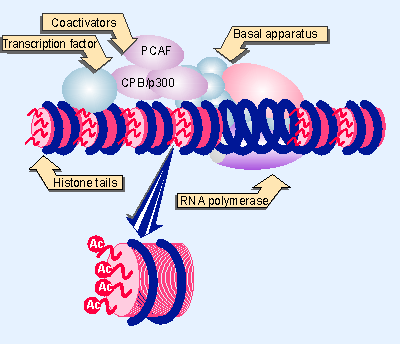 |
Figure 21.19 Coactivators may have HAT activities that acetylate the tails of nucleosomal histones. |
p300/CBP acetylates the N-terminal tails of H4 in nucleosomes. Another coactivator, called PCAF, preferentially acetylates H3 in nucleosomes. p300/CBP and PCAF form a complex that functions in transcriptional activation. In some cases yet another HAT is involved: the coactivator ACTR, which functions with hormone receptors, is itself an HAT that acts on H3 and H4, and also recruits both p300/CBP and PCAF to form a coactivating complex. One explanation for the presence of multiple HAT activities in a coactivating complex is that each HAT has a different specificity, and that multiple different acetylation events are required for activation. This enables us to redraw our picture for the action of coactivators as shown in Figure 21.19, where RNA polymerase is bound at a hypersensitive site and coactivators are acetylating histones on the nucleosomes in the vicinity (Chen et al., 1997).
The acetylases are found in large protein complexes. The TAFII145 subunit of TFIID is an acetylase. In yeast, GCN5 is part of the 1.8 MDa SAGA complex, which contains several proteins that are implicated in transcription by mutations in their genes that block initiation. Among these proteins are several TAFIIs (Grant et al., 1998). There are some functional overlaps between TFIID and SAGA, most notable that, yeast can manage with either TAFII145 or GCN5, but is damaged by the deletion of both. This suggests that an acetylase activity is essential for gene expression, but can be provided by either TFIID or SAGA (Lee et al., 2000). In both these complexes, and also some others that contains TAFFII subunits, the TAFIIs include histone-like species that probably form the basis for a structure resembling the histone octamer (Ogryzko et al., 1998).
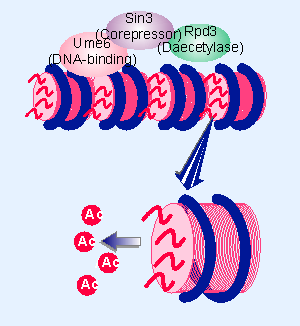 |
Figure 21.20 A repressor complex contains three components: a DNA binding subunit, a corepressor, and a histone deacetylase. |
Deacetylation is associated with repression of gene activity. In yeast, mutations in SIN3 and Rpd3 behave as though these loci repress a variety of genes. They form a complex with the DNA-binding protein Ume6, and this complex represses transcription at promoters that have the URS1 element that is bound by Ume6, as illustrated in Figure 21.20. Rpd3 has histone deacetylase activity; we do not know whether the function of Sin3 is just to bring Rpd3 to the promoter or whether it has an additional role in repression.
A similar system for repression is found in mammalian cells. The bHLH family of transcription regulators includes activators that function as heterodimers, including MyoD (see earlier). It also includes repressors, in particular the heterodimer Mad:Max, where Mad can be any one of a group of closely related proteins. The Mad:Max heterodimer (which binds to specific DNA sites) interacts with a homologue of Sin3 (called mSin3 in mouse and hSin3 in man). mSin3 is part of a repressive complex that includes histone binding proteins and the histone deacetylases HDAC1 and HDAC2. Deacetylase activity is required for repression. The modular nature of this system is emphasized by other means of employment: a corepressor (SMRT), which enables retinoid hormone receptors to repress certain target genes, functions by binding mSin3, which in turns brings the HDAC activities to the site. Another means of bringing HDAC activities to the site may be a connection with Me CP2, a protein that binds to methylated cytosines (see below).
When a coactivator acetylates DNA to assist the initiation of transcription, only the local region X~1 kb upstream and downstream of the promoter Xis affected. We do not know how this influences transcription.
Acetylation occurs at both replication (when it is transient) and at transcription (when it is maintained while the gene is active). Is it playing the same role in each case? One possibility is that the important effect is on nucleosome structure. Acetylation may be necessary to "loosen" the nucleosome core. At replication, acetylation of histones could be necessary to allow them to be incorporated into new cores more easily. At transcription, a similar effect could be necessary to allow a related change in structure, possibly even to allow the histone core to be displaced from DNA. Alternatively, acetylation could generate binding sites for other proteins that are required for transcription. In either case, deacetylation would reverse the effect.
Is the effect of acetylation quantitative or qualitative? One possibility is that a certain number of acetyl groups are required to have an effect, and the exact positions at which they occur are largely irrelevant. An alternative is that individual acetylation events have specific effects. We might interpret the existence of complexes containing multiple HAT activities in either way Xif individual enzymes have different specificities, we may need multiple activities either to acetylate a sufficient number of different positions or because the individual events are necessary for different effects upon transcription. At replication, it appears, at least with respect to histone H4, that acetylation at any two of three available positions is adequate, favoring a quantitative model in this case.
| Research | |
| Brownell, J. E. et al. (1996). Tetrahymena histone acetyltransferase A: a homologue to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843-851. | |
| Chen, H. et al. (1997). Nuclear receptor coactivator ACTR is a novel histoneacetyltransferase and forms a multimeric activation complex with P/CAF and CP/p300. Cell 90, 569-580. | |
| Grant, P. A. et al. (1998). A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94, 45-53. | |
| Lee, T. I. , Causton, H. C. , Holstege, F. C. , Shen, W. C. , Hannett, N. , Jennings, E. G. , Winston, F. , Green, M. R. , and Young, R. A. (2000). Redundant roles for the TFIID and SAGA complexes in global transcription.. Nature 405, 701-704. | |
| Ogryzko, V. V. et al. (1998). Histone-like TAFs within the PCAF histone acetylase complex. Cell 94, 35-44. | |