7. Helix-loop-helix proteins interact by combinatorial association
21.7 Helix-loop-helix proteins interact by combinatorial association |
 |
Figure 21.13 All HLH proteins have regions corresponding to helix 1 and helix 2, separated by a loop of 10-24 residues. Basic HLH proteins have a region with conserved positive charges immediately adjacent to helix 1. |
Two common features in DNA-binding proteins are the presence of helical regions that bind DNA, and the ability of the protein to dimerize. Both features are represented in the group of helix-loop-helix proteins that share a common type of sequence motif: a stretch of 40 V50 amino acids contains two amphipathic α-helices separated by a linker region (the loop) of varying length. The proteins in this group form both homodimers and heterodimers by means of interactions between the hydrophobic residues on the corresponding faces of the two helices. The helical regions are 15 V16 amino acids long, and each contains several conserved residues. Two examples are compared in Figure 21.13. The ability to form dimers resides with these amphipathic helices, and is common to all HLH proteins. The loop is probably important only for allowing the freedom for the two helical regions to interact independently of one another.
Most HLH proteins contain a region adjacent to the HLH motif itself that is highly basic, and which is needed for binding to DNA. There are ~6 conserved residues in a stretch of 15 amino acids (see Figure 21.13). Members of the group with such a region are called bHLH proteins. A dimer in which both subunits have the basic region can bind to DNA. The HLH domains probably correctly orient the two basic regions contributed by the individual subunits. We do not yet know much about the means by which HLH proteins activate transcription (Murre et al., 1989).
The bHLH proteins fall into two general groups. Class A consists of proteins that are ubiquitously expressed, including mammalian E12/E47. Class B consists of proteins that are expressed in a tissue-specific manner, including mammalian MyoD, myogenin, and Myf-5 ( a group of transcription factors that are involved in myogenesis [muscle formation]). A common modus operandi for a tissue-specific bHLH protein may be to form a heterodimer with a ubiquitous partner. There is also a group of gene products that specify development of the nervous system in D. melanogaster (where Ac-S is the tissue-specific component and da is the ubiquitous component). The Myc proteins (which are the cellular counterparts of oncogene products and are involved in growth regulation) form a separate class of bHLH proteins, whose partners and targets are different.
Dimers formed from bHLH proteins differ in their abilities to bind to DNA. For example, E47 homodimers, E12-E47 heterodimers, and MyoD-E47 heterodimers all form efficiently and bind strongly to DNA; E12 homodimerizes well but binds DNA poorly, while MyoD homodimerizes only poorly. So both dimer formation and DNA binding may represent important regulatory points. At this juncture, it is possible to define groups of HLH proteins whose members form various pairwise combinations, but not to predict from the sequences the strengths of dimer formation or DNA binding. All of the dimers in this group that bind DNA recognize the same consensus sequence, but we do not know yet whether different homodimers and heterodimers have preferences for slightly different target sites that are related to their functions.
Differences in DNA-binding result from properties of the region in or close to the HLH motif; for example, E12 differs from E47 in possessing an inhibitory region just by the basic region, which prevents DNA binding by homodimers. Some HLH proteins lack the basic region and/or contain proline residues that appear to disrupt its function. The example of the protein Id is shown in Figure 21.13. Proteins of this type have the same capacity to dimerize as bHLH proteins, but a dimer that contains one subunit of this type can no longer bind to DNA specifically. This is a forceful demonstration of the importance of doubling the DNA-binding motif in DNA-binding proteins (Benezra et al., 1990; Lassar et al., 1991; Murre et al., 1989).
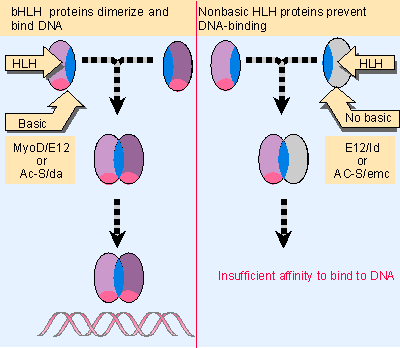 |
Figure 21.14 An HLH dimer in which both subunits are of the bHLH type can bind DNA, but a dimer in which one subunit lacks the basic region cannot bind DNA. Multiple figure |
The importance of the distinction between the nonbasic HLH and bHLH proteins is suggested by the properties of two pairs of HLH proteins: the da-Ac-S/emc pair and the MyoD/Id pair. A model for their functions in forming a regulatory network is illustrated in Figure 21.14.
In D. melanogaster, the gene emc (extramacrochaetae) is required to establish the normal spatial pattern of adult sensory organs. It functions by suppressing the functions of several genes, including da (daughterless) and the achaete-scute complex (Ac-S), which otherwise cause additional cells to take up this fate. Ac-S and da are genes of the bHLH type. The suppressor emc codes for an HLH protein that lacks the basic region. We suppose that, in the absence of emc function, the da and Ac-S proteins form dimers that activate transcription of appropriate target genes, but the production of emc protein causes the formation of heterodimers that cannot bind to DNA. So production of emc protein in the appropriate cells is necessary to suppress the function of Ac-S/da.
The formation of muscle cells is triggered by a change in the transcriptional program that requires several bHLH proteins, including MyoD. MyoD is produced specifically in myogenic cells; and, indeed, over-expression of MyoD in certain other cells can induce them to commence a myogenic program. The trigger for muscle differentiation is probably a heterodimer consisting of MyoD-E12 or MyoD-E47, rather than a MyoD homodimer. Before myogenesis begins, a member of the nonbasic HLH type, the Id protein, may bind to MyoD and/or E12 and E47 to form heterodimers that cannot bind to DNA. It binds to E12/E47 better than to MyoD, and so might function by sequestering the ubiquitous bHLH partner. Over-expression of Id can prevent myogenesis. So the removal of Id could be the trigger that releases MyoD to initiate myogenesis (Davis et al., 1987; Davis et al., 1990; for review see Weintraub, 1991).
A bHLH activator such as MyoD can be controlled in several ways. It is prevented from binding to DNA when it is sequestered by a an HLH partner such as Id. It can activate transcription when bound to bHLH partner such as E12 or E47. It can also act as a site-specific repressor when bound to another partner; the bHLH protein MyoR forms a MyoD-MyoR dimer in proliferating myoblasts that represses transcription (at the same target loci at which MyoD-E12/E47 activate transcription).
The behavior of the HLH proteins therefore illustrates two general principles of transcriptional regulation. A small number of proteins form combinatorial associations. Particular combinations have different functions with regard to DNA binding and transcriptional regulation. Differentiation may depend either on the presence or on the removal of particular partners.
| Reviews | |
| Weintraub, H. (1991). The MyoD gene family: nodal point during specification of the muscle cell lineage. Science 251, 761-766. | |
| Research | |
| Benezra, R. et al. (1990). The protein Id: a negative regulator of helix-loop-helix DNA-binding proteins. Cell 61, 49-59. | |
| Davis, R. L. et al. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987-1000. | |
| Davis, R. L. et al. (1990). The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60, 733-746. | |
| Lassar, A. B. et al. (1991). Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66, 305-315. | |
| Murre, C., McCaw, P. S., and Baltimore, D. (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777-783. | |