3. Promoter elements are defined by mutations and footprinting
20.3 Promoter elements are defined by mutations and footprinting |
| Key terms defined in this section |
| Cotransfection is the simultaneous transfection of two markers. |
Promoters have been defined in terms of their abilities to initiate transcription in suitable test systems. Having identified sequences that are needed for promoter function, we may then characterize the proteins that bind them. Several types of system have been used:
- The oocyte system follows the principles established for translation, and relies on injection of a suitable DNA template into the nucleus of the X. laevis oocyte. The RNA transcript can be recovered and analyzed. The main limitation of this system is that it is restricted to the conditions that prevail in the oocyte. It allows characterization of DNA sequences, but not of the factors that normally bind them.
- Transfection systems allow exogenous DNA to be introduced into a cultured cell and expressed. (The procedure is discussed in 17 Rearrangement of DNA.) The system is genuinely in vivo in the sense that transcription is accomplished by the same apparatus responsible for expressing the cell’s own genome. However, it differs from the natural situation because the template consists of a gene that would not usually be transcribed in the host cell. The usefulness of the system may be extended by using a variety of host cells. Cotransfection with two (or more) DNAs allows assay of the interactions between two factors.
- Transgenic systems involve the addition of a gene to the germline of an animal. Expression of the transgene can be followed in any or all of the tissues of the animal. Some common limitations apply to transgenic systems and to transfection: the additional gene often is present in multiple copies, and is integrated at a different location from the endogenous gene.
- The in vitro system takes the classic approach of purifying all the components and manipulating conditions until faithful initiation is seen. "Faithful" initiation is defined as production of an RNA starting at the site corresponding to the 5′ end of mRNA (or rRNA or tRNA precursors).
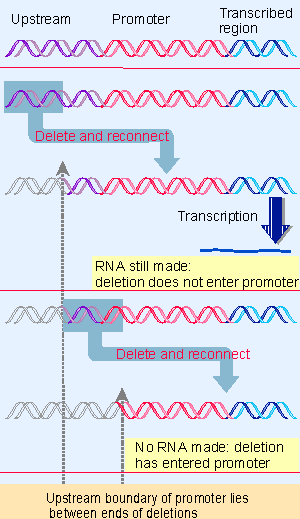 |
Figure 20.3 Promoter boundaries can be determined by making deletions that progressively remove more material from one side. When one deletion fails to prevent RNA synthesis but the next stops transcription, the boundary of the promoter must lie between them. |
We start with a particular fragment of DNA that can initiate transcription in one of these systems. Then the boundaries of the sequence constituting the promoter can be determined by reducing the length of the fragment from either end, until at some point it ceases to be active, as illustrated in Figure 20.3. The boundary upstream can be identified by progressively removing material from this end until promoter function is lost. To test the boundary downstream, it is necessary to reconnect the shortened promoter to the sequence to be transcribed (since otherwise there is no product to assay).
Several precautions are required to avoid extraneous effects. To ensure that the promoter is always in the same context, the same long upstream sequence is always placed next to it. Because termination does not occur properly in the in vitro systems, the template is cut at some distance from the promoter (usually ~500 bp downstream), to ensure that all polymerases "run off" at the same point, generating an identifiable transcript.
Once the boundaries of the promoter have been defined, the importance of particular bases within it can be determined by introducing point mutations or other rearrangements in the sequence. As with bacterial RNA polymerase, these can be characterized as up or down mutations. Some of these rearrangements affect only the rate of initiation; others influence the site at which initiation occurs, as seen in a change of the startpoint. To be sure that we are dealing with comparable products, in each case it is necessary to characterize the 5′ end of the RNA.
We can apply several criteria in identifying the sequence components of a promoter (or any other site in DNA):
- Mutations in the site prevent function in vitro or in vivo. (Many techniques now exist for introducing point mutations at particular base pairs, and in principle every position in a promoter can be mutated, and the mutant sequence tested in vitro or in vivo.)
- Proteins that act by binding to a site may be footprinted on it. There should be a correlation between the ability of mutations to prevent promoter function and to prevent binding of the factor.
- When a site recognized by a particular factor is present at multiple promoters, it should be possible to derive a consensus sequence that is bound by the factor. A new promoter should become responsive to this factor when an appropriate copy of the element is introduced.