1. Introduction
20.1 Introduction |
| Key terms defined in this section |
| Enhancer element is a cis-acting sequence that increases the utilization of (some) eukaryotic promoters, and can function in either orientation and in any location (upstream or downstream) relative to the promoter. |
Transcription in eukaryotic cells is divided into three classes. Each class is transcribed by a different RNA polymerase:
- RNA polymerase I transcribes rRNA
- RNA polymerase II transcribes mRNA
- RNA polymerase III transcribes tRNA and other small RNAs.
Accessory factors are needed for initiation, but are not required subsequently. The balance of responsibilities vis-à-vis the accessory factors is similar for all eukaryotic RNA polymerases. The factors, rather than the enzymes themselves, are principally responsible for recognizing the promoter. This contrasts with the modus operandi of bacterial RNA polymerase, in which a basic enzyme recognizes the promoters, assisted in certain cases by accessory factors.
The promoters for RNA polymerases I and II are (mostly) upstream of the startpoint, but some promoters for RNA polymerase III lie downstream of the startpoint. Each promoter contains characteristic sets of short conserved sequences that are recognized by the appropriate class of factors. RNA polymerases I and III each recognize a relatively restricted set of promoters, and rely upon a small number of accessory factors.
Promoters utilized by RNA polymerase II show more variation in sequence, and are modular in design. Short sequence elements that are recognized by transcription factors lie upstream of the startpoint. These cis-acting sites usually are spread out over a region of >200 bp. Some of these elements and the factors that recognize them are common: they are found in a variety of promoters and are used constitutively. Others are specific: they identify particular classes of genes and their use is regulated. The elements occur in different combinations in individual promoters.
The number of factors that can act in conjunction with RNA polymerase II is large. We may divide them into three general groups. We consider the first two groups in this chapter, and the third group in the next chapter:
- The general factors are required for the mechanics of initiating RNA synthesis at all promoters. They join with RNA polymerase to form a complex surrounding the startpoint, and they determine the site of initiation. The general factors together with RNA polymerase constitute the basal transcription apparatus.
- The upstream factors are DNA-binding proteins that recognize specific short consensus elements located upstream of the startpoint. The activity of these factors is not regulated; they are ubiquitous, and act upon any promoter that contains the appropriate binding site on DNA. They increase the efficiency of initiation, and are required for a promoter to function at an adequate level. The precise set of such factors that is required for full expression is characteristic of any particular promoter.
- The inducible factors function in the same general way as the upstream factors, but have a regulatory role. They are synthesized or activated at specific times or in specific tissues, and they are therefore responsible for the control of transcription patterns in time and space. The sequences that they bind are called response elements.
A promoter that contains only elements recognized by general and upstream factors should be transcribed in any cell type. Such promoters may be responsible for expression of cellular genes that are constitutively expressed (sometimes called housekeeping genes). No element/factor combination is an essential component of the promoter, which suggests that initiation by RNA polymerase II may be sponsored in many different ways. The common feature is that the upstream or inducible transcription factors bind to sequence elements located upstream of the startpoint. Binding of the factors to DNA is associated with the construction of a complex in which protein-protein interactions are important. The upstream and inducible factors function by interacting with the basal transcription apparatus, typically with certain general factors.
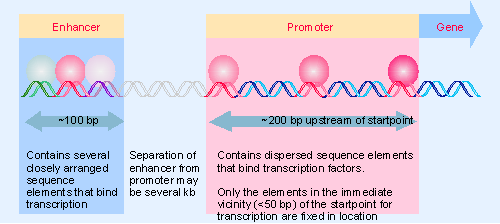 |
Figure 20.1 A typical gene transcribed by RNA polymerase II has a promoter that extends upstream from the site where transcription is initiated. The promoter contains several short (<10 bp)sequence elements that bind transcription factors, dispersed over >200 bp. An enhancer containing a more closely packed array of elements that also bind transcription factors may be located several kb distant. (DNA may be coiled or otherwise rearranged so that transcription factors at the promoter and at the enhancer interact to form a large protein complex.) |
Sequence components of the promoter are defined operationally by the demand that they must be located in the general vicinity of the startpoint and are required for initiation. The enhancer is another type of site involved in initiation. It is identified by sequences that stimulate initiation, but that are located a considerable distance from the startpoint. Enhancer elements are often targets for tissue-specific or temporal regulation. Figure 20.1 illustrates the general properties of promoters and enhancers.
The components of an enhancer resemble those of the promoter; they consist of a variety of modular elements. However, the elements are organized in a closely packed array. The elements in an enhancer function like those in the promoter, but the enhancer does not need to be near the startpoint. However, proteins bound at enhancer elements interact with proteins bound at promoter elements. The distinction between promoters and enhancers is operational, rather than implying a fundamental difference in mechanism. This view is fortified by the fact that some types of element are found in both promoters and enhancers.
Any protein that is needed for the initiation of transcription, but which is not itself part of RNA polymerase, is defined as a transcription factor. Many transcription factors act by recognizing cis-acting sites that are classified as comprising parts of promoters or enhancers. However, binding to DNA is not the only means of action for a transcription factor. A factor may recognize another factor, or may recognize RNA polymerase, or may be incorporated into an initiation complex only in the presence of several other proteins. The ultimate test for membership of the transcription apparatus is functional: a protein must be needed for transcription to occur at a specific promoter or set of promoters.
A significant difference in the transcription of eukaryotic and prokaryotic mRNAs is that initiation at a eukaryotic promoter involves a large number of factors that bind to a variety of cis-acting elements. The promoter is defined as the region containing all these binding sites, that is, which can support transcription at the normal efficiency and with the proper control. So the major feature defining the promoter for a eukaryotic mRNA is the location of binding sites for transcription factors. RNA polymerase itself binds around the startpoint, but does not directly contact the extended upstream region of the promoter. By contrast, the bacterial promoters discussed in 9 Transcription are largely defined in terms of the binding site for RNA polymerase in the immediate vicinity of the startpoint. Other sequences nearby regulate the promoter, but are generally considered distinct from it.
The common mode of regulation of eukaryotic transcription is positive: a transcription factor is provided under tissue-specific control to activate a promoter or set of promoters that contain a common target sequence. Regulation by specific repression of a target promoter is less common.
A eukaryotic transcription unit generally contains a single gene, and termination occurs beyond the end of the coding region. We should like to define the mechanism of termination, but it lacks the regulatory importance that applies in prokaryotic systems. RNA polymerases I and III terminate at discrete sequences in defined reactions, but the mode of termination by RNA polymerase II is not clear. However, the significant event in generating the 3′ end of an mRNA is not the termination event itself, but instead results from a cleavage reaction in the primary transcript (see 22 Nuclear splicing and RNA processing).
- ERP Systems Impact on Organizations
- ERP System Acquisition: A Process Model and Results From an Austrian Survey
- The Second Wave ERP Market: An Australian Viewpoint
- Intrinsic and Contextual Data Quality: The Effect of Media and Personal Involvement
- A Hybrid Clustering Technique to Improve Patient Data Quality