15. Methylation is responsible for imprinting
19.15 Methylation is responsible for imprinting |
| Key terms defined in this section |
| Imprinting describes a change in a gene that occurs during passage through the sperm or egg with the result that the paternal and maternal alleles have different properties in the very early embryo. May be caused by methylation of DNA. |
Methylation of DNA occurs at specific sites. In bacteria, it is associated with identifying the particular bacterial strain, and also with distinguishing replicated and nonreplicated DNA (see 12 The replicon). In eukaryotes, its principal known function is connected with the control of transcription. We discuss the role of methylation in controlling transcription in 21 Regulation of transcription, assuming for the present that methylation is associated with gene inactivation.
From 2 V7% of the cytosines of animal cell DNA are methylated (the value varies with the species). Most of the methyl groups are found in CG "doublets," and, in fact, the majority of the CG sequences are methylated. Usually the C residues on both strands of this short palindromic sequence are methylated, giving the structure
5′ mCpG 3′
3′ GpCm 5′
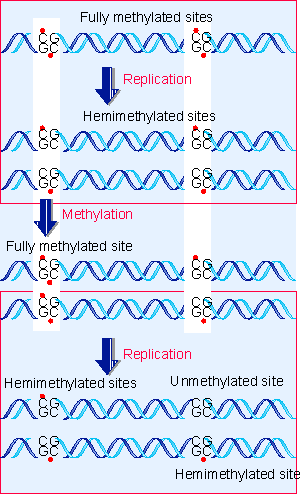 |
Figure 19.50 The state of methylated sites could be perpetuated by an enzyme that recognizes only hemimethylated sites as substrates. |
Such a site is described as fully methylated. But consider the consequences of replicating this site. Figure 19.50 shows that each daughter duplex has one methylated strand and one unmethylated strand. Such a site is called hemi-methylated (for review see Bird, 1986).
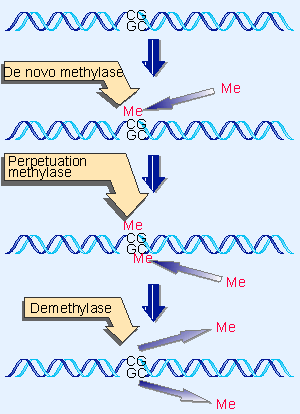 |
Figure 19.51 The state of methylation is controlled by three enzymes. |
The perpetuation of the methylated site now depends on what happens to hemimethylated DNA. If methylation of the unmethylated strand occurs, the site is restored to the fully methylated condition. However, if replication occurs next, the hemimethylated condition will be perpetuated on one daughter duplex, but the site will become unmethylated on the other daughter duplex. Figure 19.51 shows that the state of methylation of DNA is controlled by methylases, which add methyl groups to the 5 position of cytosine, and demethylases, which remove the methyl groups.
There are two types of DNA methylase, whose actions are distinguished by the state of the methylated DNA. To modify DNA at a new position requires the action of the de novo methylase, which recognizes DNA by virtue of a specific sequence. It acts only on nonmethylated DNA, to add a methyl group to one strand . There are two de novo methylases in mouse; they have different target sites, and both are essential for development (Okano et al., 1999).
A maintenance methylase acts constitutively only on hemimethylated sites to convert them to fully methylated sites. Its existence means that any methylated site is perpetuated after replication. There is one maintenance methylase in mouse, and it is essential: mouse embryos in which its gene has been disrupted do not survive past early embryogenesis (Li, Bestor, and Jaenisch, 1992).
Maintenance methylation is virtually 100% efficient, ensuring that the situation shown on the left of Figure 19.50 usually prevails in vivo. The result is that, if a de novo methylation occurs on one allele but not on the other, this difference will be perpetuated through ensuing cell divisions, maintaining a difference between the alleles that does not depend on their sequences.
The state of methylation may change at the promoters of genes that are subject to tissue-specific control. The promoters are methylated when the gene is inactive, but unmethylated when it is active.
Both methylation and demethylation occur during embryogenesis. A typical pattern of methylation is established in each sex during gametogenesis. In males, the pattern develops in two stages. Spermatocytes display the methylation pattern that is characteristic of mature sperm, so no further changes occur during spermatogenesis. But further changes are made in this pattern after fertilization. In females, the maternal pattern is imposed during oogenesis, when oocytes mature through meiosis after birth. As may be expected from the inactivity of genes in gametes, the typical state is to be methylated. However, there can be differences between the two sexes, with the result that the paternal and maternal alleles have different patterns of methylation.
A change in methylation pattern occurs during embryogenesis. All allelic differences are lost when primordial germ cells develop in the embryo; irrespective of sex, the previous patterns of methylation are erased, and a typical gene is then unmethylated. The methylation pattern of germ cells is therefore established by a two stage process: first the previous pattern is erased by a genome-wide demethylation; then the pattern specific for each sex is imposed. A major question is how the specificity of methylation is determined in the male and female gametes.
Systematic changes occur in early embryogenesis. Some sites will continue to be methylated, but others will be specifically unmethylated in cells in which a gene is expressed. From this pattern of changes, we may infer that individual sequence-specific demethylation events occur during somatic development of the organism as particular genes are activated (Chaillet et al., 1991).
The specific pattern of methyl groups in germ cells is responsible for the phenomenon of imprinting, which describes a difference in behavior between the alleles inherited from each parent. The expression of certain genes in mouse embryos depends upon the sex of the parent from which they were inherited. For example, the allele coding for IGF-II (insulin-like growth factor II) that is inherited from the father is expressed, but the allele that is inherited from the mother is not expressed. The IGF-II gene of oocytes is methylated, but the IGF-II gene of sperm is not methylated, so that the two alleles behave differently in the zygote. This is the most common pattern, but the dependence on sex is reversed for some genes. In fact, the opposite pattern (expression of maternal copy) is shown for IGF-IIR, the receptor for IGF-II (for review see Bartolomei and Tilghman, 1997).
Imprinted genes are sometimes clustered. More than half of the 17 known imprinted genes in mouse are contained in two particular regions, each containing both maternally and paternally expressed genes. This suggests the possibility that imprinting mechanisms may function over long distances. Some insights into this possibility come from deletions in the human population that cause the Prader-Willi and Angelman diseases. Most cases are caused by the same 4 Mb deletion, but the syndromes are different, depending on which parent contributed the deletion. The reason is that the deleted region contains at least one gene that is paternally imprinted and at least one that is maternally imprinted. There are some rare cases, however, with much smaller deletions. Prader-Willi syndrome can be caused by a 20 kb deletion that silences genes that are distant on either side of it. The basic effect of the deletion is to prevent a father from resetting the paternal mode to a chromosome inherited from his mother. The result is that these genes remain in maternal mode, so that the paternal as well as maternal alleles are silent in the offspring. The inverse effect is found in some small deletions that cause Angelman’s syndrome. The implication is that this region comprises some sort of "imprint center" that acts at a distance to switch one parental type to the other.
Imprinting is a classic example of epigenetic inheritance. Although the paternal and maternal alleles have identical sequences, they display different properties, depending on which parent provided them. These properties are inherited through meiosis and the subsequent somatic mitoses.
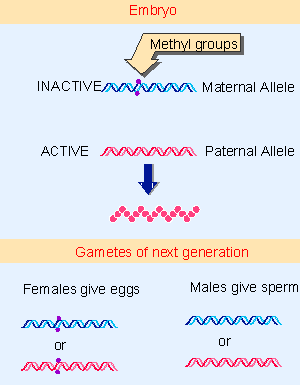 |
Figure 19.52 The parental alleles of Igf2 are differentially methylated in the early embryo, but the patterns of methylation are reset when gametes are formed by the adult. |
This sex-specific mode of inheritance requires that the pattern of methylation is established specifically during each gametogenesis. The fate of the two Igf2 alleles in a mouse is illustrated in Figure 19.52. In the early embryo, the paternal allele is nonmethylated and expressed, and the maternal allele is methylated and silent. What happens when this mouse itself forms gametes? If it is a male, the allele contributed to the sperm must be nonmethylated, irrespective of whether it was originally methylated or not. So when the maternal allele finds itself in a sperm, it must be demethylated. If the mouse is a female, the allele contributed to the egg must be methylated; so if it was originally the paternal allele, methyl groups must be added.
The behavior of Igf2, in which methylation creates an inactive imprinted state, is the most common. This reflects a direct effect of methylation on promoter activity. However, in some cases, methylation marks the active state of an imprinted gene. In these cases, the effect of methylation is indirect; for example, methylation could inactivate a silencing element.
By using an assay to detect sequences that undergo de novo methylation very early in embryonic development, a potential imprinting control site has been identified in Igf2R. It consists of an ~100 bp sequence in a CpG island in the intron of the gene. The site contains a cis-acting DNS (de novo methylation signal) element at one end that is required for denovo methylation to occur. It contains a sequence at the other end in which mutations allow the paternal allele to be methylated de novo. This suggests that this ADS (allele discriminating signal) is bound by a protein that specifically protects the paternal allele against de novo methylation. These two elements are necessary, but not sufficient, for methylation of sites in the vicinity that are differentially methylated.
This section updated 1-24-2000
| Reviews | |
| Bartolomei, M. S. and Tilghman, S. (1997). Genomic imprinting in mammals. Ann. Rev. Genet. 31, 493-525. | |
| Bird, A. P. (1986). A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Nature 321, 209-213. | |
| Research | |
| Chaillet, J. R., Vogt, T. F., Beier, D. R., and Leder, P. (1991). Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryoge. Cell 66, 77-83. | |
| Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality.. Cell 69, 915-926. | |
| Okano, M., Bell, D. W., Haber, D. A., and Li. E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development.. Cell 99, 247-257. | |