13. Heterochromatin depends on interactions with histones
19.13 Heterochromatin depends on interactions with histones |
| Key terms defined in this section |
| Epigenetic changes influence the phenotype without altering the genotype. They consist of changes in the properties of a cell that are inherited but that do not represent a change in genetic information. |
The overall properties of chromatin are influenced by interactions with proteins of the SMC (structural maintenance of chromosome) family. They are ATPases that fall into two functional groups. Condensins are involved with the control of overall structure. Cohesins are concerned with connections between sister chromatids that must be released at mitosis (see 27 Cell cycle and growth regulation).
Condensins form complexes that have a core of the heterodimer SMC2-SMC4 associated with other (non SMC) proteins. (Cohesins have a similar organization based on the heterodimeric core of SMC1-SMC3.) The condensin complex is named for its ability to cause chromatin to condense in vitro. It has an ability to introduce positive supercoils into DNA in an action that uses hydrolysis of ATP and depends on the presence of topoisomerase I. This ability is controlled by the phosphorylation of the non-SMC subunits, which occurs at mitosis. We do not know yet how this connects with other modifications of chromatin, for example, the phosphorylation of histones. The activation of the condensin complex specifically at mitosis makes it questionable whether it is also involved in the formation of interphase heterochromatin.
An interphase nucleus contains both euchromatin and heterochromatin. The condensation state of heterochromatin is close to that of mitotic chromosomes. Heterochromatin is inert. It remains condensed in interphase, is transcriptionally repressed, replicates late in S phase, and may be localized to the nuclear periphery. Centromeric heterochromatin typically consists of satellite DNAs. However, the formation of heterochromatin is not rigorously defined by sequence. When a gene is transferred, either by a chromosomal translocation or by transfection and integration, into a position adjacent to heterochromatin, it may become inactive as the result of its new location.
 |
Figure 19.44 Position effect variegation in eye color results when the white gene is integrated near heterochromatin. Cells in which white is inactive give patches of white eye, while cells in which white is active give red paPosition effect variegation in eye color results when the white gene is integrated near heterochromatin. Cells in which white is inactive give patches of white eye, while cells in which white is active give red patches. The severity of the effect is determined by the closeness of the integrated gene to heterochromatin. Photograph kindly provided by Steve Henikoff. |
Such inactivation is the result of an epigenetic effect (see also later). It may differ between individual cells in an animal, and results in the phenomenon of position effect variegation, in which genetically identical cells have different phenotypes. This has been well characterized in Drosophila. Figure 19.44 shows an example of position effect variegation in the fly eye, in which some regions lack color while others are red, because the white gene is inactivated by adjacent heterochromatin in some cells, while it remained active in other cells.
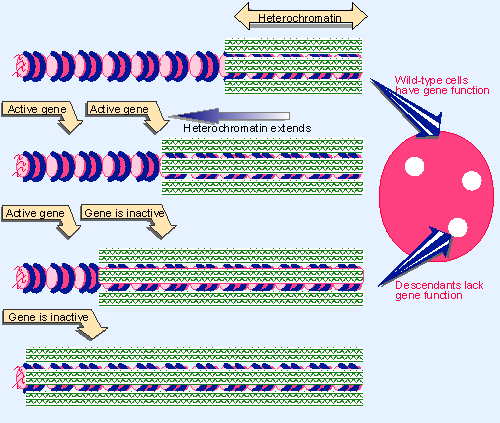 |
Figure 19.45 Extension of heterochromatin inactivates genes. The probability that a gene will be inactivated depends on its distance from the heterochromatin region. |
The explanation for this effect is shown in Figure 19.45. Inactivation spreads from heterochromatin into the adjacent region for a variable distance. In some cells it goes far enough to inactivate a nearby gene, but in others it does not. This happens at a certain point in embryonic development, and after that point the state of the gene is inherited by all the progeny cells. Cells descended from an ancestor in which the gene was inactivated form patches corresponding to the phenotype of loss-of-function (in the case of white, absence of color).
The closer a gene lies to heterochromatin, the higher the probability that it will be inactivated. This suggests that the formation of heterochromatin may be a two-stage process: a nucleation event occurs at a specific sequence; and then the inactive structure propagates along the chromatin fiber. The distance for which the inactive structure extends is not precisely determined (for example, by boundary elements) and may be stochastic, being influenced by parameters such as the quantities of limiting protein components.
Genes that are closer to heterochromatin are more likely to be inactivated, and will therefore be inactive in a greater proportion of cells. On this model, the boundaries of a heterochromatic region might be terminated by exhausting the supply of one of the proteins that is required.
The effect of telomeric silencing in yeast is analogous to position effect variegation in Drosophila; genes translocated to a telomeric location show the same sort of variable loss of activity. This results from a spreading effect that propagates from the telomeres.
A second form of silencing occurs in yeast. Yeast mating type is determined by the activity of a single active locus (MAT), but the genome contains two other copies of the mating type sequences (HML and HMR), which are maintained in an inactive form. We discuss the contribution of these loci to yeast mating type in 17 Rearrangement of DNA. The silent loci HML and HMR share many properties with heterochromatin, and could be regarded as constituting regions of heterochromatin in miniature.
The existence of a common basis for both types of silencing in yeast is suggested by their common reliance on the same set of genetic loci. Mutations in any one of a number of genes cause HML and HMR to become activated, and also relieve the inactivation of genes that have been integrated near telomeric heterochromatin. The products of these loci therefore function to maintain the inactive state of both types of heterochromatin (for review see Loo and Rine, 1995).
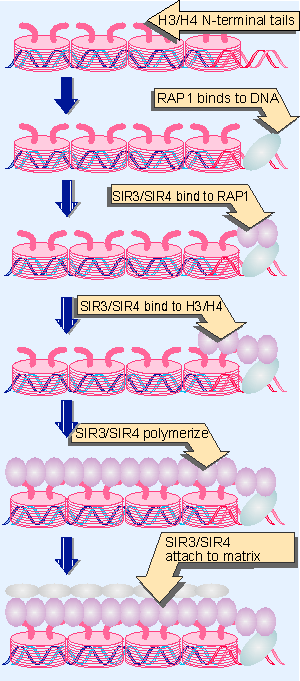 |
Figure 19.46 Formation of heterochromatin is initiated when RAP1 binds to DNA. SIR3/4 bind to RAP1 and also to histones H3/H4. The complex polymerizes along chromatin and may connect telomeres to the nuclear matrix. |
Figure 19.46 proposes a model for actions of these proteins. Only one of them is a sequence-specific DNA-binding protein. This is RAP1, which binds to the C1-3A repeats at the telomeres, and also binds to the cis-acting silencer elements that are needed for repression of HML and HMR. The proteins SIR3 and SIR4 interact with RAP1 and also with one another (they may function as a heteromultimer). SIR3/SIR4 interact with the N-terminal tails of the histones H3 and H4. (In fact, the first evidence that histones might be involved directly in formation of heterochromatin was provided by the discovery that mutations abolishing silencing at HML/HMR map to genes coding for H3 and H4 (Shore and Nasmyth, 1987; Kayne et al., 1988; for review see Thompson and Hecht, 1993).
RAP1 has the crucial role of identifying the DNA sequences at which heterochromatin forms. It recruits SIR3/SIR4, and they interact directly with the histones H3/H4. Once SIR3/SIR4 have bound to histones H3/H4, the complex may polymerize further, and spread along the chromatin fiber. This may inactivate the region, either because coating with SIR3/SIR4 itself has an inhibitory effect, or because binding to histones H3/H4 induces some further change in structure. We do not know what limits the spreading of the complex. The C-terminus of SIR3 has a similarity to nuclear lamin proteins (constituents of the nuclear matrix) and may be responsible for tethering heterochromatin to the nuclear periphery (Palladino et al., 1993; Moretti et al., 1994; Hecht et al., 1995).
Another specialized chromatin structure forms at the centromere. Its nature is suggested by the properties of an S. cerevisiae mutation, cse4, that disrupts the structure of the centromere. Cse4p is a protein that is related to histone H3. A mammalian centromeric protein, CENP-A, has a related sequence. Genetic interactions between cse4 and CDE-II, and between cse4 and a mutation in the H4 histone gene, suggest that a histone octamer may form around a core of Cse4p-H4, and then the centromeric complexes CBF1 and CBF3 may attach to form the centromere (Bloom and Carbon, 1982; Meluh et al., 1998).
| Reviews | |
| Loo, S. and Rine, J. (1995). Silencing and heritable domains of gene expression. Ann. Rev. Cell Dev. Biol. 11, 519-548. | |
| Thompson, J. S., Hecht, A., and Grunstein, M. (1993). Histones and the regulation of heterochromatin in yeast. Cold Spring Harbor Symp. Quant. Biol. 58, 247-256. | |
| Research | |
| Bloom, K. S. and Carbon, J. (1982). Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29, 305-317. | |
| Hecht, A., Laroche, T., Strahl-Bolsinger, S., Gasser, S. M., and Grunstein, M. (1995). Histone H3 and H4 N-termini interact with the silent information regulators SIR3 and SIR4: a molecular model for the formation of heterochromatin in yeast. Cell 80, 583-592. | |
| Kayne, P. S., Kim, U. J., Han. M., Mullen, R. J., Yoshizaki, F., and Grunstein, M. (1988). Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55, 27-39. | |
| Meluh, P. B. et al. (1998). Cse4p is a component of the core centromere of S. cerevisiae. Cell 94, 607-613. | |
| Moretti, P., Freeman, K., Coodly, L., and Shore, D. (1994). Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8, 2257-2269. | |
| Palladino, F., Laroche, T., Gilson, E., Axelrod, A., Pillus, L., and Gasser, S. M. (1993). SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75, 543-555. | |
| Shore, D. and Nasmyth, K. (1987). Purification and cloning of a DNA-binding protein from yeast that binds to both silencer and activator elements. Cell 51, 721-732. | |