9. TnA transposition requires transposase and resolvase
15.9 TnA transposition requires transposase and resolvase |
Replicative transposition is the only mode of mobility of the TnA family, which consists of large (~5 kb) transposons. They are not composites relying on IS-type transposition modules, but comprise independent units carrying genes for transposition as well as for features such as drug resistance. The TnA family includes several related transposons, of which Tn3 and Tn1000 (formerly called γδ) are the best characterized. They have the usual terminal feature of closely related inverted repeats, generally ~38 bp in length. Cis-acting deletions in either repeat prevent transposition of an element. A 5 bp direct repeat is generated at the target site. They carry resistance markers such as ampr.
The two stages of TnA-mediated transposition are accomplished by the transposase and the resolvase, whose genes, tnpA and tnpR, are identified by recessive mutations. The transposition stage involves the ends of the element, as it does in IS Vtype elements. Resolution requires a specific internal site, a feature unique to the TnA family (for review see Sherratt, 1989).
Mutants in tnpA cannot transpose. The gene product is a transposase that binds to a sequence of ~25 bp located within the 38 bp of the inverted terminal repeat. A binding site for the E. coli protein IHF exists adjacent to the transposase binding site; and transposase and IHF bind cooperatively. The transposase recognizes the ends of the element and also makes the staggered 5 bp breaks in target DNA where the transposon is to be inserted. IHF is a DNA-binding protein that is often involved in assembling large structures in E. coli; its role in the transposition reaction may not be essential.
The tnpR gene product has dual functions. It acts as a repressor of gene expression and it provides the resolvase function.
Mutations in tnpR increase the transposition frequency. The reason is that TnpR represses the transcription of both tnpA and its own gene. So inactivation of TnpR protein allows increased synthesis of TnpA, which results in an increased frequency of transposition. This implies that the amount of the TnpA transposase must be a limiting factor in transposition.
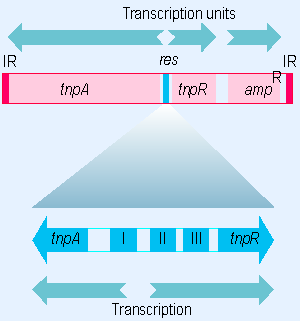 |
Figure 15.18 Transposons of the TnA family have inverted terminal repeats, an internal res site, and three known genes. |
The tnpA and tnpR genes are expressed divergently from an A PT-rich intercistronic control region, indicated in the map of Tn3 given in Figure 15.18. Both effects of TnpR are mediated by its binding in this region.
In its capacity as the resolvase, TnpR is involved in recombination between the direct repeats of Tn3 in a cointegrate structure. A cointegrate can in principle be resolved by a homologous recombination between any corresponding pair of points in the two copies of the transposon. But the Tn3 resolution reaction occurs only at a specific site (Droge et al., 1990).
The site of resolution is called res. It is identified by cis-acting deletions that block completion of transposition, causing the accumulation of cointegrates. In the absence of res, the resolution reaction can be substituted by RecA-mediated general recombination, but this is much less efficient.
The sites bound by the TnpR resolvase are summarized in the lower part of Figure 15.18. Binding occurs independently at each of three sites, each 30 V40 bp long. The three binding sites share a sequence homology that defines a consensus sequence with dyad symmetry (Grindley et al., 1982).
Site I includes the region genetically defined as the res site; in its absence, the resolution reaction does not proceed at all. However, resolution also involves binding at sites II and III, since the reaction proceeds only poorly if either of these sites is deleted. Site I overlaps with the startpoint for tnpA transcription. Site II overlaps with the startpoint for tnpR transcription; an operator mutation maps just at the left end of the site.
Do the sites interact? One possibility is that binding at all three sites is required to hold the DNA in an appropriate topology. Binding at a single set of sites may repress tnpA and tnpR transcription without introducing any change in the DNA.
An in vitro resolution assay uses a cointegrate Vlike DNA molecule as substrate. The substrate must be supercoiled; its resolution produces two catenated circles, each containing one res site. The reaction requires large amounts of the TnpR resolvase; no host factors are needed. Resolution occurs in a large nucleoprotein structure. Resolvase binds to each res site, and then the bound sites are brought together to form a structure ~10 nm in diameter. Changes in supercoiling occur during the reaction, and DNA is bent at the res sites by the binding of transposase.
Resolution is a nonreplicative reaction; bonds are broken and rejoined without demand for input of energy. The products identify an intermediate stage in cointegrate resolution; they consist of resolvase covalently attached to both 5′ ends of double Vstranded cuts made at the res site. The cleavage occurs symmetrically at a short palindromic region to generate two base extensions. Expanding the view of the crossover region located in site I, we can describe the cutting reaction as:
5′ T T A T A A 3′
3′ A A T A T T 5′
5′ T T A T + protein-A A 3′
3′ A A-protein T A T T 5′
The reaction resembles the action of lambda Int at the att sites (see 14 Recombination and repair). Indeed, 15 of the 20 bp of the res site are identical to the bases at corresponding positions in att. This suggests that the site-specific recombination of lambda and resolution of TnA have evolved from a common type of recombination reaction; and indeed, we shall see in 24 Immune diversity that recombination involving immunoglobulin genes has the same basis.
The reactions themselves are analogous in terms of manipulation of DNA, although resolution occurs only between intramolecular sites, whereas the recombination between att sites is intermolecular and directional (as seen by the differences in attB and attP sites). However, the mechanism of protein action is different in each case. Resolvase functions in a manner in which four subunits bind to the recombining res sites. Each subunit makes a single-strand cleavage. Then a reorganization of the subunits relative to one another physically moves the DNA strands, placing them in a recombined conformation. Then the nicks can be sealed.
| Reviews | |
| Sherratt, D. (1989). Tn3 and related transposable elements: site-specific recombination and transposition. In Mobile DNA, Eds. Berg, D. E. and Howe, M. American Society of Microbiology, Washington DC 163-185. | |
| Research | |
| Droge, P. et al. (1990). The two functional domains of gamma delta resolvase act on the same recombination site: implications for the mechanism of strand exchange. Proc. Nat. Acad. Sci. USA 87, 5336-5340. | |
| Grindley, N. D. et al. (1982). Transposon-mediated site-specific recombination: identification of three binding sites for resolvase at the res sites of gd and Tn3. Cell 30, 19-27. | |