2. Insertion sequences are simple transposition modules
15.2 Insertion sequences are simple transposition modules |
| Key terms defined in this section |
| Direct repeats are identical (or related) sequences present in two or more copies in the same orientation in the same molecule of DNA; they are not necessarily adjacent. Inverted terminal repeats are the short related or identical sequences present in reverse orientation at the ends of some transposons. IS is an abbreviation for insertion sequence Transposase is the enzyme activity involved in insertion of transposon at a new site. |
Transposable elements were first identified in the form of spontaneous insertions in bacterial operons. Such an insertion prevents transcription and/or translation of the gene in which it is inserted. Many different types of transposable elements have now been characterized (for review see Kleckner, 1977; Calos and Miller, 1980; Kleckner, 1981; Berg and Howe, 1989).
The simplest transposons are called insertion sequences (reflecting the way in which they were detected). Each type is given the prefix IS, followed by a number that identifies the type. (The original classes were numbered IS1 V4; later classes have numbers reflecting the history of their isolation, but not corresponding to the total number of elements so far isolated!)
The IS elements are normal constituents of bacterial chromosomes and plasmids. A standard strain of E. coli is likely to contain several (<10) copies of any one of the more common IS elements. To describe an insertion into a particular site, a double colon is used; so λ::IS1 describes an IS1 element inserted into phage lambda.
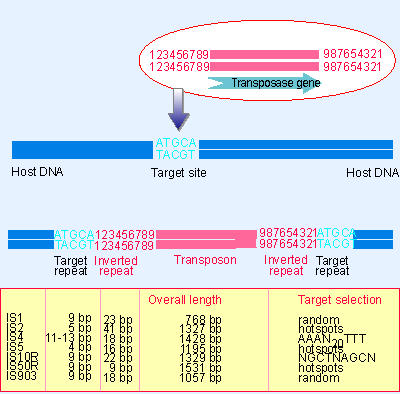 |
Figure 15.1 Transposons have inverted terminal repeats and generate direct repeats of flanking DNA at the target site. In this example, the target is a 5 bp sequence. The ends of the transposon consist of inverted repeats of 9 bp, where the numbers 1 through 9 indicate a sequence of base pairs. |
The IS elements are autonomous units, each of which codes only for the proteins needed to sponsor its own transposition. Each IS element is different in sequence, but there are some common features in organization. The structure of a generic transposon before and after insertion at a target site is illustrated in Figure 15.1, which also summarizes the details of some common IS elements (for review see Galas and Chandler, 1989).
An IS element ends in short inverted terminal repeats; usually the two copies of the repeat are closely related rather than identical. As illustrated in the figure, the presence of the inverted terminal repeats means that the same sequence is encountered proceeding toward the element from the flanking DNA on either side of it.
When an IS element transposes, a sequence of host DNA at the site of insertion is duplicated. The nature of the duplication is revealed by comparing the sequence of the target site before and after an insertion has occurred. Figure 15.1 shows that at the site of insertion, the IS DNA is always flanked by very short direct repeats. (In this context, "direct" indicates that two copies of a sequence are repeated in the same orientation, not that the repeats are adjacent.) But in the original gene (prior to insertion), the target site has the sequence of only one of these repeats. In the figure, the target site consists of the sequence ATGCATACGT. After transposition, one copy of this sequence is present on either side of the transposon (Grindley, 1978; Johnsrud et al., 1978).
The sequence of the direct repeat varies among individual transposition events undertaken by a transposon, but the length is constant for any particular IS element (a reflection of the mechanism of transposition). The most common length for the direct repeats is 9 bp.
An IS element therefore displays a characteristic structure in which its ends are identified by the inverted terminal repeats, while the adjacent ends of the flanking host DNA are identified by the short direct repeats. When observed in a sequence of DNA, this type of organization is taken to be diagnostic of a transposon, and makes a prima facie case that the sequence originated in a transposition event.
Most IS elements insert at a variety of sites within host DNA. However, some show (varying degrees of) preference for particular hotspots (for review see Craig, 1997).
The inverted repeats define the ends of a transposon. Recognition of the ends is common to transposition events sponsored by all types of transposon. Cis-acting mutations that prevent transposition are located in the ends, which are recognized by a protein(s) responsible for transposition. The protein is called a transposase.
All the IS elements except IS1 contain a single long coding region, starting just inside the inverted repeat at one end, and terminating just before or within the inverted repeat at the other end. This codes for the transposase. IS1 has a more complex organization, with two separate reading frames; the transposase is produced by making a frameshift during translation to allow both reading frames to be used.
The frequency of transposition varies among different elements. The overall rate of transposition is ~10 V3 V10 V4 per element per generation. Insertions in individual targets occur at a level comparable with the spontaneous mutation rate, usually ~10 V5 V10 V7 per generation. Reversion (by precise excision of the IS element) is usually infrequent, with a range of rates of 10 V6 to 10 V10 per generation, ~103 times less frequent than insertion.
| Reviews | |
| Berg, D. E. and Howe, M. (1989). Mobile DNA. American Society for Microbiology, Washington DC. | |
| Berg, D. E. and Howe, M. (1989). Mobile DNA. American Society for Microbiology, Washington DC. | |
| Calos, M. and Miller, J. H. (1980). Transposable elements. Cell 20, 579-595. | |
| Craig, N. L. (1997). Target site selection in transposition. Ann. Rev. Biochem 66, 437-474. | |
| Galas, D. J. and Chandler, M. (1989). Bacterial insertion sequences. In Mobile DNA, Eds. Berg, D. E. and Howe, M. American Society of Microbiology, Washington DC 109-162. | |
| Kleckner, N. (1977). Translocatable elements in prokaryotes. Cell 11, 11-23. | |
| Kleckner, N. (1981). Transposable elements in prokaryotes. Ann. Rev. Genet. 15, 341-404. | |
| Research | |
| Grindley, N. D. (1978). IS1 insertion generates duplication of a 9 bp sequence at its target site. Cell 13, 419-426. | |
| Johnsrud, L., Calos, M. P., and Miller, J. H. (1978). The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell 15, 1209-1219. | |