8. Rolling circles produce multimers of a replicon
12.8 Rolling circles produce multimers of a replicon |
| Key terms defined in this section |
| Rolling circle is a mode of replication in which a replication fork proceeds around a circular template for an indefinite number of revolutions; the DNA strand newly synthesized in each revolution displaces the strand synthesized in the previous revolution, giving a tail containing a linear series of sequences complementary to the circular template strand. |
The structures generated by replication depend on the relationship between the template and the replication fork. The critical features are whether the template is circular or linear, and whether the replication fork is engaged in synthesizing both strands of DNA or only one.
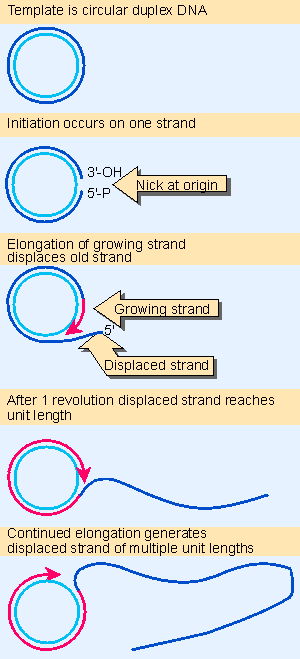 |
Figure 12.16 The rolling circle generates a multimeric single-stranded tail. Animated figure |
Replication of only one strand is used to generate copies of some circular molecules. A nick opens one strand, and then the free 3′ VOH end generated by the nick is extended by the DNA polymerase. The newly synthesized strand displaces the original parental strand. The ensuing events are depicted in Figure 12.16.
This type of structure is called a rolling circle, because the growing point can be envisaged as rolling around the circular template strand. It could in principle continue to do so indefinitely. As it moves, the replication fork extends the outer strand and displaces the previous partner (Gilbert and Dressler, 1968).
Because the newly synthesized material is covalently linked to the original material, the displaced strand has the original unit genome at its 5′ end. The original unit is followed by any number of unit genomes, synthesized by continuing revolutions of the template. Each revolution displaces the material synthesized in the previous cycle.
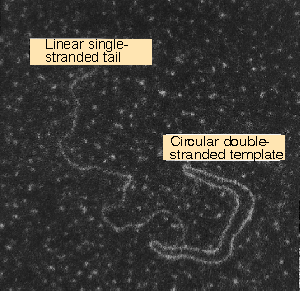 |
Figure 12.17 A rolling circle appears as a circular molecule with a linear tail by electron microscopy. Photograph kindly provided by David Dressler. |
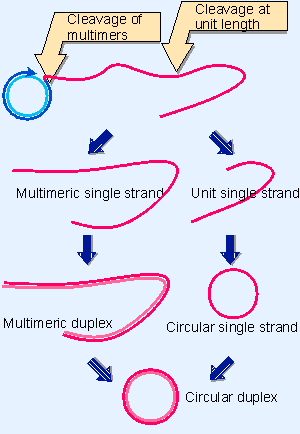 |
Figure 12.18 Rolling circles can be used for varying purposes, depending on the fate of the displaced tail. Cleavage at unit length generates monomers, which can be converted to duplex and circular forms. Cleavage of multimers generates a series of tandemly repeated copies of the original unit. Note that the conversion to double-stranded form could occur earlier, before the tail is cleaved from the rolling circle. |
An example is shown in the electron micrograph of Figure 12.17. The rolling circle is put to several uses in vivo. Some pathways that are used to replicate DNA are depicted in Figure 12.18.
Cleavage of a unit length tail generates a copy of the original circular replicon in linear form. The linear form may be maintained as a single strand or may be converted into a duplex by synthesis of the complementary strand (which is identical in sequence to the template strand of the original rolling circle).
The rolling circle provides a means for amplifying the original (unit) replicon. This mechanism is used to generate amplified rDNA in the Xenopus oocyte. The genes for rRNA are organized as a large number of contiguous repeats in the genome. A single repeating unit from the genome is converted into a rolling circle. The displaced tail, containing many units, is converted into duplex DNA; later it is cleaved from the circle so that the two ends can be joined together to generate a large circle of amplified rDNA. The amplified material therefore consists of a large number of identical repeating units.
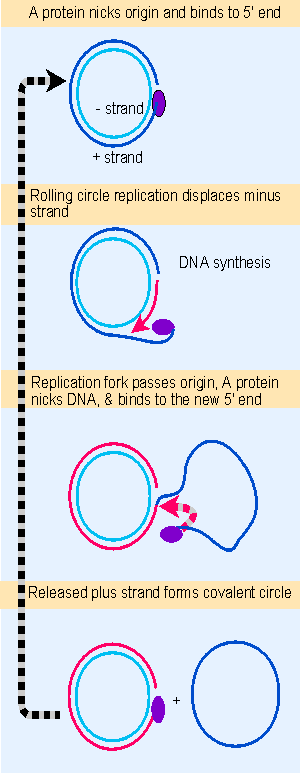 |
Figure 12.19 fX174 RF DNA is a template for synthesizing single-stranded viral circles. The A protein remains attached to the same genome through indefinite revolutions, each time nicking the origin on the viral (+) strand and transferring to the new 5 F end. At the same time, the released viral strand is circularized. Animated figure |
Replication by rolling circles is common among bacteriophages. Unit genomes can be cleaved from the displaced tail, generating monomers that can be packaged into phage particles or used for further replication cycles. A more detailed view of a phage replication cycle that is centered on the rolling circle is given in Figure 12.19. Phage φdurham consists of a single-stranded circular DNA, known as the plus (+) strand. A complementary strand, called the minus ( V ) strand, is synthesized. This action generates the duplex circle shown at the top of the figure, which is then replicated by a rolling circle mechanism.
The duplex circle is converted to a covalently closed form, which becomes supercoiled. A protein coded by the phage genome, the A protein, nicks the (+) strand of the duplex DNA at a specific site that defines the origin for replication. After nicking the origin, the A protein remains connected to the 5′ end that it generates, while the 3′ end is extended by DNA polymerase.
The structure of the DNA plays an important role in this reaction, for the DNA can be nicked only when it is supercoiled. The A protein is able to bind to a single-stranded decamer fragment of DNA that surrounds the site of the nick. This suggests that the supercoiling is needed to assist the formation of a single-stranded region that provides the A protein with its binding site. (An enzymatic activity in which a protein cleaves duplex DNA and binds to a released 5′ end is sometimes called a relaxase.) The nick generates a 3′ VOH end and a 5′ Vphosphate end (covalently attached to the A protein), both of which have roles to play in φX174 replication.
Using the rolling circle, the 3′ VOH end of the nick is extended into a new chain. The chain is elongated around the circular ( V) strand template, until it reaches the starting point and displaces the origin. Now the A protein functions again. It remains connected with the rolling circle as well as to the 5′ end of the displaced tail, and it is therefore in the vicinity as the growing point returns past the origin. So the same A protein is available again to recognize the origin and nick it, now attaching to the end generated by the new nick. The cycle can be repeated indefinitely.
Following this nicking event, the displaced single (+) strand is freed as a circle. The A protein is involved in the circularization. In fact, the joining of the 3′ and 5′ ends of the (+) strand product is accomplished by the A protein as part of the reaction by which it is released at the end of one cycle of replication, and starts another cycle.
The A protein has an unusual property that may be connected with these activities. It is cis-acting in vivo. (This behavior is not reproduced in vitro, as can be seen from its activity on any DNA template in a cell-free system.) The implication is that in vivo the A protein synthesized by a particular genome can attach only to the DNA of that genome. We do not know how this is accomplished. However, its activity in vitro shows how it remains associated with the same parental ( V) strand template. The A protein has two active sites; this may allow it to cleave the "new" origin while still retaining the "old" origin; then it ligates the displaced strand into a circle.
The displaced (+) strand may follow either of two fates after circularization. During the replication phase of viral infection, it may be used as a template to synthesize the complementary ( V) strand. The duplex circle may then be used as a rolling circle to generate more progeny. During phage morphogenesis, the displaced (+) strand is packaged into the phage virion.
| Research | |
| Gilbert, W. and Dressler, D. (1968). DNA replication: the rolling circle model. Cold Spring Harbor Symp. Quant. Biol. 33, 473-484. | |