4. Each eukaryotic chromosome contains many replicons
12.4 Each eukaryotic chromosome contains many replicons |
| Key terms defined in this section |
| S phase is the restricted part of the eukaryotic cell cycle during which synthesis of DNA occurs. |
In eukaryotic cells, the replication of DNA is confined to part of the cell cycle. S phase usually lasts a few hours in a higher eukaryotic cell. Replication of the large amount of DNA contained in a eukaryotic chromosome is accomplished by dividing it into many individual replicons. Only some of these replicons are engaged in replication at any point in S phase. Presumably each replicon is activated at a specific time during S phase, although the evidence on this issue is not decisive (for review see Fangman and Brewer, 1991).
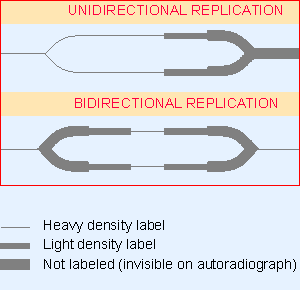 |
Figure 12.5 Different densities of radioactive labeling can be used to distinguish unidirectional and bidirectional replication. |
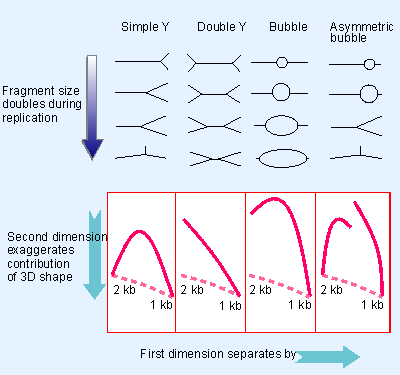 |
Figure 12.6 The position of the origin and the number of replicating forks determine the shape of a replicating restriction fragment, which can be followed by its electrophoretic path (solid line). The dashed line shows the path for a linear DNA. |
The start of S phase is signaled by the activation of the first replicons. Over the next few hours, initiation events occur at other replicons in an ordered manner. Much of our knowledge about the properties of the individual replicons is derived from autoradiographic studies, generally using the types of protocols illustrated in Figure 12.5 and Figure 12.6. Chromosomal replicons usually display bidirectional replication.
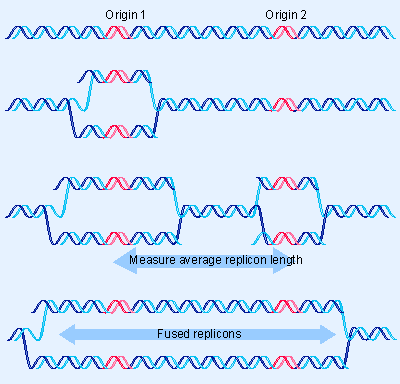 |
Figure 12.8 Measuring the size of the replicon requires a stretch of DNA in which adjacent replicons are active. |
How large is the average replicon, and how many are there in the genome? A difficulty in characterizing the individual unit is that adjacent replicons may fuse to give large replicated eyes, as illustrated in Figure 12.8. The approach usually used to distinguish individual replicons from fused eyes is to rely on stretches of DNA in which several replicons can be seen to be active, presumably captured at a stage when all have initiated around the same time, but before the forks of adjacent units have met.
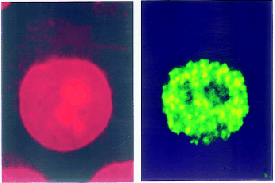 |
Figure 12.9 Replication forks are organized into foci in the nucleus. Cells were labeled with BrdU. The leftmost panel was stained with propidium iodide to identify bulk DNA. The right panel was stained using an antibody to BrdU to identify replicating DNA. Photographs kindly provided by A. D. Mills and Ron Laskey. |
"Regional" controls might produce this sort of activation pattern, in which groups of replicons are initiated more or less coordinately, as opposed to a mechanism in which individual replicons are activated one by one in dispersed areas of the genome. Two structural features suggest the possibility of large-scale organization. Quite large regions of the chromosome can be characterized as "early replicating" or "late replicating," implying that there is little interspersion of replicons that fire at early or late times. And visualization of replicating forks by labeling with DNA precursors identifies 100 V300 "foci" instead of uniform staining; each focus shown in Figure 12.9 probably contains >300 replication forks. The foci could represent fixed structures through which replicating DNA must move.
In groups of active replicons, the average size of the unit is measured by the distance between the origins (that is, between the midpoints of adjacent replicons). The rate at which the replication fork moves can be estimated from the maximum distance that the autoradiographic tracks travel during a given time.
Individual replicons in eukaryotic genomes are relatively small, typically ~40 kb in yeast or fly, ~100 kb in animals cells. However, they can vary >10-fold in length within a genome. The rate of replication is ~2000 bp/min, which is much slower than the 50,000 bp/min of bacterial replication fork movement.
From the speed of replication, it is evident that a mammalian genome could be replicated in ~1 hour if all replicons functioned simultaneously. But S phase actually lasts for >6 hours in a typical somatic cell, which implies that no more than 15% of the replicons are likely to be active at any given moment. (There are some exceptional cases, such as the early embryonic divisions of Drosophila embryos, where the duration of S phase is compressed by the simultaneous functioning of a large number of replicons. (Blumenthal et al., 1974))
How are origins selected for initiation at different times during S phase? In S. cerevisiae, the default appears to be for origins to replicate early, but cis-acting sequences can cause origins linked to them to replicate at late times.
Available evidence suggests that chromosomal replicons do not have termini at which the replication forks cease movement and (presumably) dissociate from the DNA. It seems more likely that a replication fork continues from its origin until it meets a fork proceeding toward it from the adjacent replicon. We have already mentioned the potential topological problem of joining the newly synthesized DNA at the junction of the replication forks.
| Reviews | |
| Fangman, W. L. and Brewer, B. J. (1991). Activation of replication origins within yeast chromosomes. Ann. Rev. Cell Biol. 7, 375-402. | |
| Research | |
| Blumenthal, A. B., Kriegstein, H. J., and Hogness, D. S. (1974). The units of DNA replication inD. melanogaster chromosomes. Cold Spring Harbor Symp. Quant. Biol. 38, 205-223. | |