10. Adverse growth conditions provoke the stringent response
10.10 Adverse growth conditions provoke the stringent response |
| Key terms defined in this section |
| Idling reaction is the production of pppGpp and ppGpp by ribosomes when an uncharged tRNA is present in the A site; triggers the stringent response. Stringent response refers to the ability of a bacterium to shut down synthesis of tRNA and ribosomes in a poor-growth medium. |
When bacteria find themselves in such poor growth conditions that they lack a sufficient supply of amino acids to sustain protein synthesis, they shut down a wide range of activities. This is called the stringent response. We can view it as a mechanism for surviving hard times: the bacterium husbands its resources by engaging in only the minimum of activities until nutrient conditions improve, when it reverses the response and again engages its full range of metabolic activities.
The stringent response causes a massive (10 V20 ) reduction in the synthesis of rRNA and tRNA. This alone is sufficient to reduce the total amount of RNA synthesis to only 5 V10% of its previous level. The synthesis of certain mRNAs is reduced, leading to an overall reduction of ~3 in mRNA synthesis. The rate of protein degradation is increased. Many metabolic adjustments occur, as seen in reduced synthesis of nucleotides, carbohydrates, lipids, etc.
The stringent response causes the accumulation of two unusual nucleotides. ppGpp is guanosine tetraphosphate, with diphosphates attached to both 5′ and 3′ positions. pppGpp is guanosine pentaphosphate, with a 5′ triphosphate group and a 3′ diphosphate. These nucleotides are typical small-molecule effectors that function by binding to target proteins to alter their activities. Sometimes they are known collectively as (p)ppGpp (Cashel and Gallant, 1969; for review see Cashel and Rudd, 1987).
(p)ppGpp functions to regulate coordinately a large number of cellular activities. Its production is controlled in two ways. A drastic increase in (p)ppGpp is triggered by the stringent response. And there is also a general inverse correlation between (p)ppGpp levels and the bacterial growth rate, which is controlled by some unknown means.
Deprivation of any one amino acid, or mutation to inactivate any aminoacyl-tRNA synthetase, is sufficient to initiate the stringent response. The trigger that sets the entire series of events in train is the presence of uncharged tRNA in the A site of the ribosome. Under normal conditions, of course, only aminoacyl-tRNA is placed in the A site by EF-Tu (see 6 Protein synthesis). But when there is no aminoacyl-tRNA available to respond to a particular codon, the uncharged tRNA becomes able to gain entry. Of course, this blocks any further progress by the ribosome; and it triggers an idling reaction.
The components involved in producing (p)pGpp via the idling reaction have been identified through the existence of relaxed (rel) mutants. rel mutations abolish the stringent response, so that starvation for amino acids does not cause any reduction in stable RNA synthesis or alter any of the other reactions that are usually seen.
The most common site of relaxed mutation lies in the gene relA, which codes for a protein called the stringent factor. This factor is associated with the ribosomes, although the amount is rather low Xsay, <1 molecule for every 200 ribosomes. So perhaps only a minority of the ribosomes are able to produce the stringent response.
Ribosomes obtained from stringent bacteria can synthesize ppGpp and pppGpp in vitro, provided that the A site is occupied by an uncharged tRNA specifically responding to the codon. Ribosomes extracted from relaxed mutants cannot perform this reaction; but they are able to do so if the stringent factor is added.
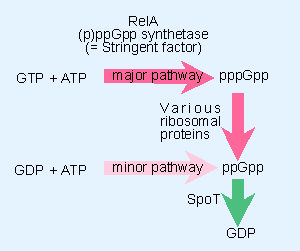 |
Figure 10.28 Stringent factor catalyzes the synthesis of pppGpp and ppGpp; ribosomal proteins can dephosphorylate pppGpp to ppGpp. |
Figure 10.28 shows the pathways for synthesis of (p)ppGpp. The stringent factor (RelA) is an enzyme that catalyzes the synthetic reaction in which ATP is used to donate a pyrophosphate group to the 3′ position of either GTP or GDP. The formal name for this activity is (p)ppGpp synthetase.
How is ppGpp removed when conditions return to normal? A gene called spoT codes for an enzyme that provides the major catalyst for ppGpp degradation. The activity of this enzyme causes ppGpp to be rapidly degraded, with a half-life of ~20 sec; so the stringent response is reversed rapidly when synthesis of (p)ppGpp ceases. spoT mutants have elevated levels of ppGpp, and grow more slowly as a result.
The RelA enzyme uses GTP as substrate more frequently, so that pppGpp is the predominant product. However, pppGpp is converted to ppGpp by several enzymes; among those able to perform this dephosphorylation are the translation factors EF-Tu and EF-G. The production of ppGpp via pppGpp is the most common route, and ppGpp is the usual effector of the stringent response.
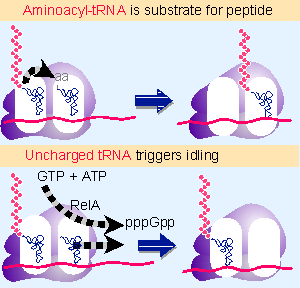 |
Figure 10.29 In normal protein synthesis, the presence of aminoacyl-tRNA in the A site is a signal for peptidyl transferase to transfer the polypeptide chain, followed by movement catalyzed by EF-G; but under stringent conditions, the presence of uncharged tRNA causes RelA protein to synthesize (p)ppGpp and to expel the tRNA. |
The response of the ribosome to entry of uncharged tRNA is compared with normal protein synthesis in Figure 10.29. When EF-Tu places aminoacyl-tRNA in the A site, peptide bond synthesis is followed by ribosomal movement. But when uncharged tRNA is paired with the codon in the A site, the ribosome remains stationary and engages in the idling reaction (Haseltine and Block, 1973).
How does the state of the ribosome control the activity of RelA enzyme? An indication of the nature of the interaction is revealed by relaxed mutations in another locus, originally called relC, which turns out to be the same as rplK, which codes for the 50S subunit protein L11. This protein is located in the vicinity of the A and P sites, in a position to respond to the presence of a properly paired but uncharged tRNA in the A site. A conformational change in this protein or some other component could activate the RelA enzyme, so that the idling reaction occurs instead of polypeptide transfer from the peptidyl-tRNA.
One round of (p)ppGpp synthesis is associated with release of the uncharged tRNA from the A site, so that synthesis of (p)ppGpp is a continuing response to the level of uncharged tRNA. So under limiting conditions, a ribosome stalls when no aminoacyl-tRNA is available to respond to the codon in the A site. Entry of uncharged tRNA triggers the synthesis of a (p)ppGpp molecule, and the resulting expulsion of the uncharged tRNA allows the situation to be reassessed. Depending upon the availability of aminoacyl-tRNA, the ribosome resumes polypeptide synthesis or undertakes another idling reaction.
What does ppGpp do? It is an effector for controlling several reactions, including the inhibition of transcription. Many effects have been reported, among which two stand out:
- Initiation of transcription is specifically inhibited at the promoters of operons coding for rRNA. Mutations of stringently regulated promoters can abolish stringent control, which suggests that the effect requires an interaction with specific promoter sequences.
- The elongation phase of transcription of many or most templates is reduced by ppGpp. The cause is increased pausing by RNA polymerase. This effect is responsible for the general reduction in transcription efficiency when ppGpp is added in vitro.
The use of ppGpp is just one aspect of a more general regulatory network that relates production of ribosomes to the growth rate. The level of protein synthesis increases in proportion with the growth rate. This is accomplished by increasing the production of ribosomes as cells grow more rapidly. The cell therefore needs some general indicator of growth rate that can be used to control the synthesis of ribosomes. The indicator appears to be NTP levels, and the target for their action is the control of transcription of rRNA.
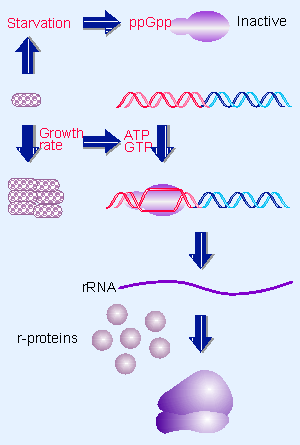 |
Figure 10.30 Nucleotide levels control initiation of rRNA transcription. |
Figure 10.30 summarizes the systems that are used to control rRNA transcription in response to growth rate. Under conditions of starvation, ppGpp is produced, and (among its various actions) inhibits initiation at the promoters of the rrn loci that code for rRNA. As growth rate increases, the levels of ATP and GTP increase. These increase the rate of initiation at the rrn promoters (for review see Condon et al., 1995).
The rrn promoters in E. coli form atypical open complexes with RNA polymerase. The open complexes are unusually unstable. The result is that the main factor governing the rate of initiation becomes the decay rate of the open complex. Increased concentration of the initiating nucleotide (which is ATP at six of the rrn promoters and GTP at the seventh) drives the initiation reaction forward by stabilizing the open complex.
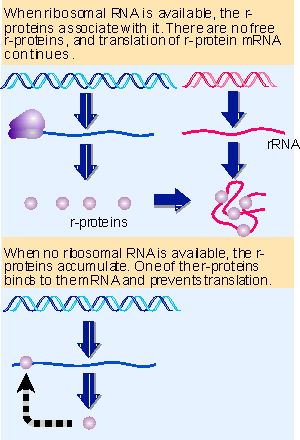 |
Figure 10.35 Translation of the r-protein operons is autogenously controlled and responds to the level of rRNA. Animated figure |
The level of rRNA controls the production of ribosomes (by a feedback loop in which free [excess] rRNA inhibits ribosomal protein synthesis, as we see in the next section in Figure 10.35). This means that the production of ribosomes, and thus of protein synthesis, in turn respond to the levels of ATP and GTP, which reflect the nutritional condition of the cell. So concentrations of particular nucleotides control ribosome synthesis in response to normal changes in growth rate and the more extreme conditions of starvation.
| Reviews | |
| Cashel, M. and Rudd, K. E. (1987). The stringent response In E. coli and S. typhimurium. In E. coli and S. typhimurium, Ed. F. C. Neidhardt, American Society for Microbiology, Washington DC 1410-1429. | |
| Condon, C., Squires, C., and Squires, C. L. (1995). Control of rRNA transcription in E. coli. Microbiol. Rev. 59, 623-645. | |
| Research | |
| Cashel, M. and Gallant, J. (1969). Two compounds implicated in the function of the RC gene of E. coli. Nature 221, 838-841. | |
| Haseltine, W. A. and Block, R. (1973). Synthesis of guanosine tetra and pentaphosphate requires the presence of a codon specific uncharged tRNA in the acceptor site of ribosomes. Proc. Nat. Acad. Sci. USA 70, 1564-1568. | |