11. Antitermination depends on specific sites
9.11 Antitermination depends on specific sites |
| Key terms defined in this section |
| Antitermination proteins allow RNA polymerase to transcribe through certain terminator sites. |
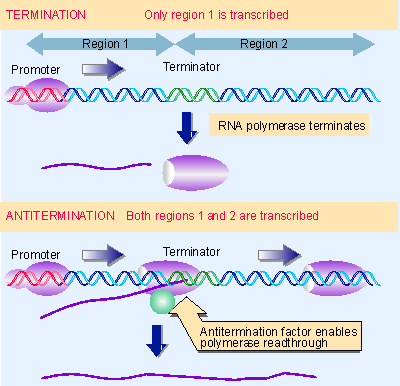 |
Figure 9.30 Antitermination can be used to control transcription by determining whether RNA polymerase terminates or reads through a particular terminator into the following region. |
Antitermination is used as a control mechanism in both phage regulatory circuits and bacterial operons. Figure 9.30 shows that antitermination controls the ability of the enzyme to read past a terminator into genes lying beyond. In the example shown in the figure, the antitermination factor regulates the expression of region 2.
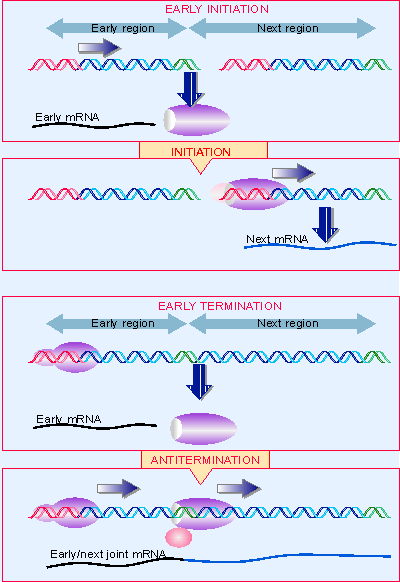 |
Figure 9.31 Switches in transcriptional specificity can be controlled at initiation or termination. Multiple figure |
Antitermination was discovered in bacteriophage infections. A common feature in the control of phage infection is that very few of the phage genes (the "early" genes) can be transcribed by the bacterial host RNA polymerase. Among these genes, however, are regulator(s) whose product(s) allow the next set of phage genes to be expressed. Two common types of action for such a regulator protein are to sponsor initiation at new (phage) promoters or to cause the host polymerase to read through phage terminators. Figure 9.31 compares the use of new promoters as a control mechanism with the use of antitermination.
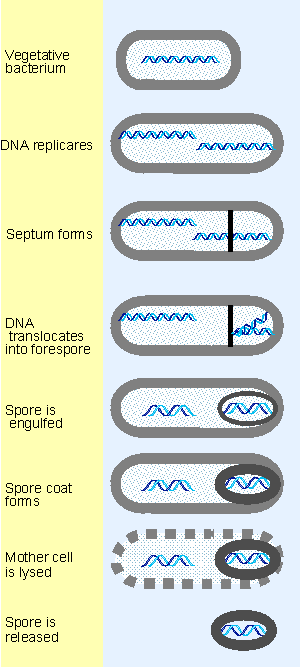 |
Figure 9.22 Sporulation involves the differentiation of a vegetative bacterium into a mother cell that is lysed and a spore that is released. |
One mechanism for recognizing new phage promoters is to replace the sigma factor of the host enzyme with another factor that redirects its specificity in initiation (see Figure 9.22 and 9.24). An alternative mechanism is to synthesize a new phage RNA polymerase. In either case, the critical feature that distinguishes the new set of genes is their possession of different promoters from those originally recognized by host RNA polymerase. The two sets of transcripts are independent; as a consequence, early gene expression can cease after the new sigma factor or polymerase has been produced.
Antitermination provides an alternative mechanism for phages to control the switch from early genes to the next stage of expression. The use of antitermination depends on a particular arrangement of genes. The early genes lie adjacent to the genes that are to be expressed next, but are separated from them by terminator sites. If termination is prevented at these sites, the polymerase reads through into the genes on the other side. So in antitermination, the same promoters continue to be recognized by RNA polymerase. So the new genes are expressed only by extending the RNA chain to form molecules that contain the early gene sequences at the 5′ end and the new gene sequences at the 3′ end. Since the two types of sequence remain linked, early gene expression inevitably continues (for review see 102).
The best characterized example of antitermination is provided by phage lambda, with which the phenomenon was discovered. The host RNA polymerase initially transcribes two genes, which are called the immediate early genes. The transition to the next stage of expression is controlled by preventing termination at the ends of the immediate early genes, with the result that the delayed early genes are expressed. (We discuss the overall regulation of lambda development in more detail in the next chapter.)
The regulator gene that controls the switch from immediate early to delayed early expression is identified by mutations in lambda gene N that can transcribe only the immediate early genes; they proceed no further into the infective cycle. The same effect is seen when gene 28 of phage SPO1 is mutated to prevent the production of σ gp28. From the genetic point of view, the mechanisms of new initiation and antitermination are similar. Both are positive controls in which an early gene product must be made by the phage in order to express the next set of genes.
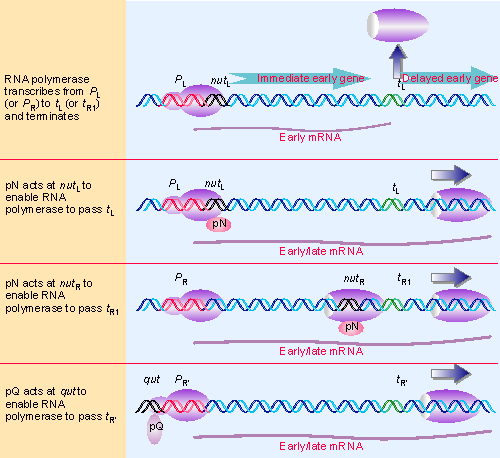 |
Figure 9.32 Host RNA polymerase transcribes lambda genes and terminates at t sites. pN allows it to read through terminators in the L and R1 units; pQ allows it to read through the R F terminator. The sites at which pN acts (nut) and at which pQ acts (qut) are located at different relative positions in the transcription units. |
Antitermination is the mechanism used by phage lambda to control the transition out of the early stage. It is summarized in Figure 9.32. There are two transcription units of immediate early genes, which are transcribed from the promoters PL and PR. Transcription by E. coli RNA polymerase itself stops at the terminators tL1 and tR1, respectively. Both terminators depend on rho; in fact, these were the terminators with which rho was originally identified. The situation is changed by expression of the N gene. The product pN is an antitermination protein that allows RNA polymerase to read through tL1 and tR1 into the delayed early genes beyond them.
Like other phages, still another control is needed to express the late genes that code for the components of the phage particle. This switch is regulated by gene Q, itself one of the delayed early genes. Its product, pQ, is another antitermination protein, one that specifically allows RNA polymerase initiating at another site, the late promoter PR′, to read through a terminator that lies between it and the late genes. So by employing antitermination proteins with different specificities, a cascade for gene expression can be constructed.
The different specificities of pN and pQ establish an important general principle: RNA polymerase interacts with transcription units in such a way that an ancillary factor can sponsor antitermination specifically for some transcripts. Termination can be controlled with the same sort of precision as initiation. What sites are involved in controlling the specificity of termination?
The antitermination activity of pN is highly specific, but the antitermination event is not determined by the terminators tL1 and tR1; the recognition site needed for antitermination lies upstream in the transcription unit, that is, at a different place from the terminator site at which the action eventually is accomplished. This conclusion establishes a general principle. When we know the site on DNA at which some protein exercises its effect, we cannot assume that this coincides with the DNA sequence that it initially recognizes. They may be separate.
The recognition sites required for pN action are called nut (for N utilization). The sites responsible for determining leftward and rightward antitermination are described as nutL and nutR, respectively. Mapping of nut mutations locates nutL between the startpoint of PL and the beginning of the N coding region; and nutR lies between the end of the cro gene and tL1. This means that the two nut sites lie in different positions relative to the organization of their transcription units. Whereas nutL is near the promoter, nutR is near to the terminator. (qut is different yet again, and lies within the promoter.)
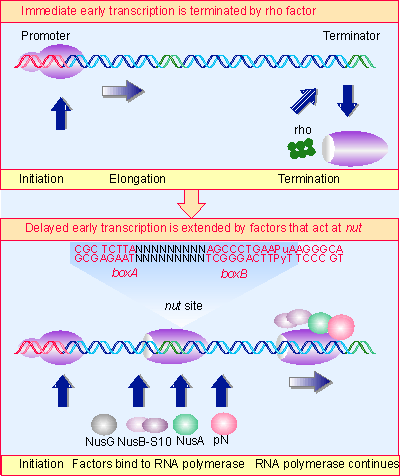 |
Figure 9.33 Ancillary factors bind to RNA polymerase as it passes certain sites. The nut site consists of two sequences. NusB-S10 join core enzyme as it passes boxA. Then NusA and pN protein bind as polymerase passes boxB. The presence of pN allows the enzyme to read through the terminator, producing a joint mRNA that contains immediate early sequences joined to delayed early sequences. Multiple figure |
How does antitermination occur? When pN recognizes the nut site, it must act on RNA polymerase to ensure that the enzyme can no longer respond to the terminator. The variable locations of the nut sites indicate that this event is linked neither to initiation nor to termination, but can occur to RNA polymerase as it elongates the RNA chain past the nut site. As illustrated in Figure 9.33, the polymerase then becomes a juggernaut that continues past the terminator, heedless of its signal. (This reaction involves antitermination at rho-dependent terminators, but pN also suppresses termination at intrinsic terminators.)
Is the ability of pN to recognize a short sequence within the transcription unit an example of a more widely used mechanism for antitermination? Other phages, related to lambda, have different N genes and different antitermination specificities. The region of the phage genome in which the nut sites lie has a different sequence in each of these phages, and each phage must therefore have characteristic nut sites recognized specifically by its own pN. Each of these pN products must have the same general ability to interact with the transcription apparatus in an antitermination capacity, but has a different specificity for the sequence of DNA that activates the mechanism.
pN and pQ both affect the ability of core RNA polymerase to terminate, but they interact differently with the enzyme. pN interacts with the core enzyme as it passes particular DNA sites, but pQ interacts with the holoenzyme during the initiation phase. In fact, σ70 is required for the interaction with pQ. This reinforces the view of RNA polymerase as an interactive structure in which conformational changes induced at one phase may affect its activity at a later phase.
| Reviews | |
| 102: | Greenblatt, J., Nodwell, J. R., and Mason, S. W. (1993). Transcriptional antitermination. Nature 364, 401-406. |