1.9 Architectures and Adaptive Procedures
|
1.9 Architectures and Adaptive Procedures
The tabletop prototype discussed in the previous section can be thought of as an extreme abstraction of the recognition-action dynamics of a biological cell. The cell is crudely pictured as a mixing chamber. The syringes roughly correspond to receptors that serve to introduce signaling substances into the chamber. The enzyme is the primary processing component, acting on the medium to trigger an output signal that could potentially control an action.
As noted above, more enzymes and signaling substances could be added. Alternative designs are possible—for example, designs with enzymes that are embedded in a matrix in an ordered way. The potential nonadditivity of the superposed response surface increases, thereby increasing the complexity of the transformation. The goal is to create a repertoire of high-complexity basis functions for implementing input-output transforms that cannot be accommodated by programmable architectures (as discussed in section 1.2).
Three issues arise: how to migrate the tabletop prototype to a chip, how to generate a useful repertoire of transformations, and how to use these chips as molecular coprocessors for a conventional architecture or to organize them into novel architectural designs.
Current advances in lab-on-a-chip technology open up a number of possible migration pathways. Figure 1.13 visualizes one of these (Zauner and Conrad 1997). This coprocessor comprises two layers; a molecular layer that contains the macromolecules and milieu components, and an optoelectronic layer that serves as the input-output interface. The molecular layer could be a sealed fluid film, gel matrix, or Langmuir-Blodgett film (Blodgett 1935). Proteins could be embedded in the film and materials moved around using microfluidic techniques (Hadd et al. 1997; Chohen et al. 1999; Unger et al. 2000). Specific molecular components are selected to couple the molecular layer to the optoelectronic layer for input and output. A pattern of light signals introduces the pattern to be classified. The induced pattern of milieu features is then fused by the conformational dynamics of the embedded proteins.
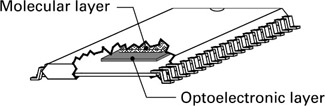
Figure 1.13: Hypothetical molecular coprocessor combining microfluidics and integrated optoelectronics. (Reproduced with permission from Optical Memory and Neural Networks 1997, 6: 157–173. 1997 Allerton Press, Inc.)
The resulting conformation change produces spectroscopically identifiable signals, either directly or indirectly through catalytic change in the concentration of a lightabsorbing substance. The optoelectronic layer would include integrated optics (e.g., waveguides, gratings) for coupling to the molecular layer and could incorporate integrated circuits for interfacing with a conventional electronic environment. Activities of multiple proteins in the molecular layer could be used for readout, but this depends on spectrophotometers with parallel capabilities in an appropriate wavelength range to come on-line. The choice of parameters for readout of the dynamics constitutes the interpretation.
The second issue concerns the adaptation of the physical dynamics and the interpretation. The tuning of our tabletop prototype was done by varying the substances used for coding of the inputs and essentially by ad hoc variation of the substrate concentration. A response surface was then constructed that could be used to elicit different functionalities, attention being focused in the present case on the twovariable logic functions (because only two input lines were used). The number of signal substances could be increased. The number of enzyme species included could be increased and their type varied. New macromolecular species could be evolved with specific capabilities—using, for example, protein-engineering techniques (Beaudry and Joyce 1992; Gao et al. 1997). The combinatorics clearly grows explosively, as it does in natural biological evolution. Response-surface methodology (Box and Draper 1987) can be used to prune this gigantic search space. The surfaces would be explored for features that could provide useful input-output transformations and the next steps of variation could be focused on the most interesting regions of the surface. The whole process can be automated.
The technology is available for this development program, but the evolution of suitable transforms must, of course, be a long-term, continuing process. As a first step, we envisage the development of a limited class of modules that can serve as molecular coprocessors for conventional machines. These could be used as preprocessors to transform complex input patterns into rigidly defined output patterns that can be rapidly processed by digital techniques. The conventional architecture would provide the procedural capabilities, but these would be complemented and synergized by the self-organizing dynamics of the molecular coprocessors.
As more molecular basis functions become available, it should be possible to build up an architecture with a more neuromolecular character. Artificial neural networks are essentially built up out of a set of fairly simple transforms. The situation in the brain is arguably quite different. The neuronal units exhibit a diversity of capabilities that draw on internal molecular dynamics. Complex interweavings of self-organization and procedural processes mediate what, according to our earlier considerations, are the high complexity programs that cannot be accommodated by conventional architectures.
Our group has developed a virtual system, referred to as the artificial neuromolecular (ANM) architecture, along this line (Chen 1993). The system consists of neurons controlled by an internal signal integration mechanism modeled after the neuronal cytoskeleton. Read-in elements represent molecules of the input layer in a molecular chip; readout elements correspond to molecules that trigger output firing. Neurons fire when a locus occupied by a readout element is sufficiently activated. The input-output transform performed by the neuron is adapted by varying internal parameters (read-in locations, readout locations, structure of the signal integration network) and varying the connections to other neurons. A repertoire of specialpurpose transforms is thus created. Memory manipulation mechanisms that are essentially procedural in nature are then used to orchestrate the different neuron types into assemblages capable of executing yet higher complexity transforms, again using a variation-selection evolutionary technique.
The ANM architecture has been applied to a variety of 64-bit pattern-recognition problems (the input interface being currently limited in this way). These include maze navigation (Chen and Conrad 1994), Chinese character recognition (Chen and Conrad 1997), and most recently, hepatitis diagnosis (Chen 2000). The power of the system lies in its computational adaptability properties. It is a virtual system run on top of a conventional base machine. It uses the limited resources of a low-complexity machine to achieve computational adaptability, but this must be at the expense of other desirable features that programs using the same resources differently might exhibit. The molecular processing in the neurons—particularly the readout—is, of course, nominal. The readouts are just threshold elements. It would be too computationally costly to simulate the conformational dynamics that allows contextsensitive fusion of milieu features. The reasonable supposition is that implementing the architecture with real molecules would enormously increase the complexity of the programs that it is capable of embodying, thereby affording concomitant expansion of the problem domains that it is capable of managing.
|