8. Elongation factor T loads aminoacyl-tRNA into the A site
6.8 Elongation factor T loads aminoacyl-tRNA into the A site |
| Key terms defined in this section |
| Elongation factors (EF in prokaryotes, eEF in eukaryotes) are proteins that associate with ribosomes cyclically, during addition of each amino acid to the polypeptide chain. |
Once the complete ribosome is formed at the initiation codon, the stage is set for a cycle in which aminoacyl-tRNA enters the A site of a ribosome whose P site is occupied by peptidyl-tRNA. Any aminoacyl-tRNA except the initiator can enter the A site. Its entry is mediated by an elongation factor (EF-Tu in bacteria). The process is similar in eukaryotes.
Just like its counterpart in initiation (IF-2), this factor, EF-Tu, is associated with the ribosome only during its sponsorship of aminoacyl-tRNA entry. Once the aminoacyl-tRNA is in place, EF-Tu leaves the ribosome, to work again with another aminoacyl-tRNA. So it displays the cyclic association with, and dissociation from, the ribosome that is the hallmark of the accessory factors.
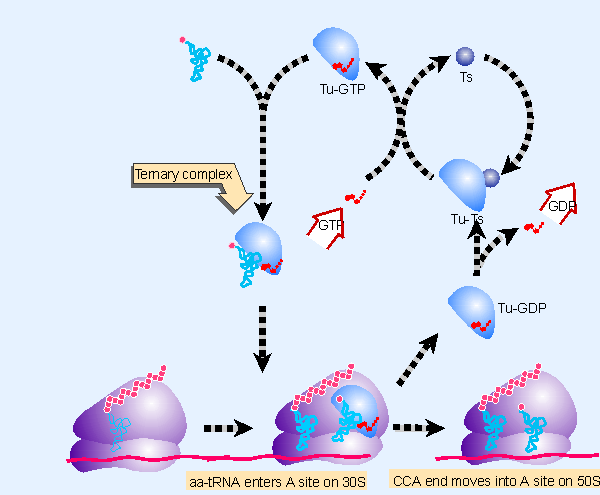 |
Figure 6.20 EF-Tu-GTP places aminoacyl-tRNA on the ribosome and then is released as EF-Tu-GDP. EF-Ts is required to mediate the replacement of GDP by GTP. The reaction consumes GTP and releases GDP. The only aminoacyl-tRNA that cannot be recognized by EF-Tu-GTP is fMet-tRNAf, whose failure to bind prevents it from responding to internal AUG or GUG codons. |
The pathway for aminoacyl-tRNA entry to the A site is illustrated in Figure 6.20. EF-Tu carries a guanine nucleotide. This factor is a monomeric G protein whose activity is controlled by the state of the guanine nucleotide (see introduction on G proteins):
- When GTP is present, the factor is in its active state.
- When the GTP is hydrolyzed to GDP, the factor becomes inactive.
- Activity is restored when the GDP is replaced by GTP.
The binary complex of EF-Tu PGTP binds aminoacyl-tRNA to form a ternary complex of aminoacyl-tRNA PEF-Tu PGTP. The ternary complex binds only to the A site of ribosomes whose P site is already occupied by peptidyl-tRNA. This is the critical reaction in ensuring that the aminoacyl-tRNA and peptidyl-tRNA are correctly positioned for peptide bond formation.
Aminoacyl-tRNA is loaded into the A site in two stages. First the anticodon end binds to the A site of the 30S subunit. Then codon-anticodon recognition triggers a change in the conformation of EF-Tu. The GTP is cleaved. The CCA end of the tRNA now moves into the A site on the 50S subunit. The binary complex EF-Tu PGDP is released. This form of EF-Tu is inactive and does not bind aminoacyl-tRNA effectively.
Another factor, EF-Ts, mediates the regeneration of the used form, EF-Tu PGDP, into the active form, EF-Tu PGTP. First, EF-Ts displaces the GDP from EF-Tu, forming the combined factor EF-Tu PEF-Ts. Then the EF-Ts is in turn displaced by GTP, reforming EF-Tu PGTP. The active binary complex binds aminoacyl-tRNA; and the released EF-Ts can recycle.
There are ~70,000 molecules of EF-Tu per bacterium (~5% of the total bacterial protein), which approaches the number of aminoacyl-tRNA molecules. This implies that most aminoacyl-tRNAs are likely to be present in ternary complexes. There are only ~10,000 molecules of EF-Ts per cell (about the same as the number of ribosomes).
The role of GTP in the ternary complex has been studied by substituting an analog that cannot be hydrolyzed. The compound GMP-PCP has a methylene bridge in place of the oxygen that links the β and γ phosphates in GTP. In the presence of GMP-PCP, a ternary complex can be formed that binds aminoacyl-tRNA to the ribosome. But the peptide bond cannot be formed. So the presence of GTP is needed for aminoacyl-tRNA to be bound at the A site; but the hydrolysis is not required until later.
Kirromycin is an antibiotic that inhibits the function of EF-Tu. When EF-Tu is bound by kirromycin, it remains able to bind aminoacyl-tRNA to the A site. But the EF-Tu PGDP complex cannot be released from the ribosome. Its continued presence prevents formation of the peptide bond between the peptidyl-tRNA and the aminoacyl-tRNA. As a result, the ribosome becomes "stalled" on mRNA, bringing protein synthesis to a halt.
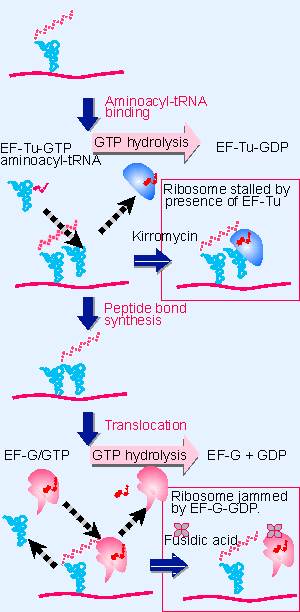 |
Figure 6.24 Binding of factors EF-Tu and EF-G alternates as ribosomes accept new aminoacyl-tRNA, form peptide bonds, and translocate. |
This effect of kirromycin demonstrates that inhibiting one step in protein synthesis blocks the next step. The reason is that the continued presence of EF-Tu prevents the aminoacyl end of aminoacyl-tRNA from entering the A site on the 50S subunit (see Figure 6.24). So the release of EF-Tu PGDP is needed for the ribosome to undertake peptide bond formation. The same principle is seen at other stages of protein synthesis: one reaction must be completed properly before the next can occur.
The interaction with EF-Tu also plays a role in quality control. Aminoacyl-tRNAs are brought into the A site without knowing whether their anticodons will fit the codon. The hydrolysis of EF-Tu PGTP is relatively slow: because it takes longer than the time required for an incorrect aminoacyl-tRNA to dissociate from the A site, most incorrect species are removed at this stage. The release of EF-Tu PGDP after hydrolysis also is slow, so any surviving incorrect aminoacyl-tRNAs may dissociate at this stage. The basic principle is that the reactions involving EF-Tu occur slowly enough to allow incorrect aminoacyl-tRNAs to dissociate before they become trapped in protein synthesis.
In eukaryotes, the factor eEF-1α is responsible for bringing aminoacyl-tRNA to the ribosome, again in a reaction that involves cleavage of a high-energy bond in GTP. It is homologous to its prokaryotic counterpart (EF-Tu), and similarly is an abundant protein. After hydrolysis of GTP, the active form is regenerated by the factor eEF-1βγ, a counterpart to EF-Ts.