4. DNA replication is semiconservative
1.4 DNA replication is semiconservative |
| Key terms defined in this section |
| DNA polymerases are enzymes that synthesize a daughter strand(s) of DNA (under direction from a DNA template). May be involved in repair or replication. DNAases are enzymes that attack bonds in DNA. Endonucleases cleave bonds within a nucleic acid chain; they may be specific for RNA or for single-stranded or double-stranded DNA. Exonucleases cleave nucleotides one at a time from the end of a polynucleotide chain; they may be specific for either the 5´ or 3´ end of DNA or RNA. Parental strands of DNA are the two complementary strands of duplex DNA before replication. Replication fork is the point at which strands of parental duplex DNA are separated so that replication can proceed. Ribonucleases are enzymes that degrade RNA. Exo(ribo)nucleases work progressively, typically degrading one base at a time from the 3´ end toward the 5 ´ end. Endo(ribo)nucleases make single cuts within the RNA chain. RNA polymerases are enzymes that synthesize RNA using a DNA template (formally described as DNA-dependent RNA polymerases). RNAases are enzymes that degrade RNA. Semiconservative replication is accomplished by separation of the strands of a parental duplex, each then acting as a template for synthesis of a complementary strand. |
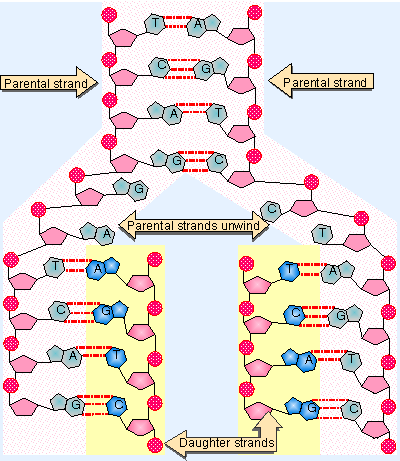 |
Figure 1.9 Base pairing provides the mechanism for replicating DNA. |
It is crucial that the genetic material is reproduced accurately. Because the two polynucleotide strands are joined only by hydrogen bonds, they are able to separate without requiring breakage of covalent bonds. The specificity of base pairing suggests that each of the separated parental strands could act as a template for the synthesis of a complementary daughter strand, as depicted in Figure 1.9. The principle is that a new daughter strand is assembled on each parental strand. The sequence of the daughter strand is dictated by the parental strand; an A in the parental strand causes a T to be placed in the daughter strand, a parental G directs incorporation of a daughter C, and so on.
The top part of the figure shows a parental duplex and the lower part shows the two daughter duplexes that are being produced by complementary base pairing. The (unreplicated) parental duplex consists of the original two parental strands. Replication requires the two parental strands to be separated so that each can be used as a template for synthesis of a complement. Each of the daughter duplexes is identical in sequence with the original parent, and contains one parental strand and one newly synthesized strand. The structure of DNA carries the information needed to perpetuate its sequence.
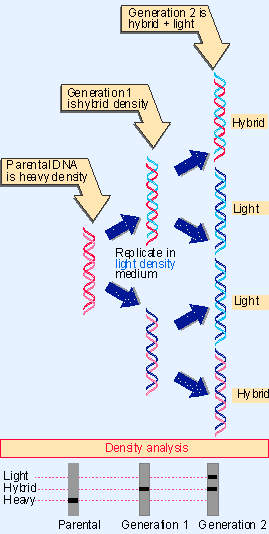 |
Figure 1.10 Replication of DNA is semiconservative. |
The consequences of this mode of replication are illustrated for the DNA molecule as a whole in Figure 1.10. The parental duplex is replicated to form two daughter duplexes, each of which consists of one parental strand and one (newly synthesized) daughter strand. The unit conserved from one generation to the next is one of the two individual strands comprising the parental duplex. This behavior is called semiconservative replication.
The figure illustrates a prediction of this model. If the parental DNA carries a "heavy" density label because the organism has been grown in medium containing a suitable isotope (such as 15N), its strands can be distinguished from those that are synthesized when the organism is transferred to a medium containing normal "light" isotopes.
The parental DNA consists of a duplex of two heavy strands (red). After one generation of growth in light medium, the duplex DNA is "hybrid" in density Xit consists of one heavy parental strand (red) and one light daughter strand (blue). After a second generation, the two strands of each hybrid duplex have separated; each gains a light partner, so that now half of the duplex DNA remains hybrid while half is entirely light (both strands are blue).
The individual strands of these duplexes are entirely heavy or entirely light. This pattern was confirmed experimentally in the Meselson-Stahl experiment of 1958, which followed the semiconservative replication of DNA through three generations of growth of E. coli. When DNA was extracted from bacteria and its density measured by centrifugation, the DNA formed bands corresponding to its density Xheavy for parental, hybrid for the first generation, and half hybrid and half light in the second generation (Meselson and Stahl, 1958).
Replication involves a major disruption of the structure of DNA. However, although the two strands of the parental duplex must separate, they do not exist as single strands. The disruption of structure is only transient and is reversed as the daughter duplex is formed. So only a small part of the DNA loses the duplex structure at any moment.
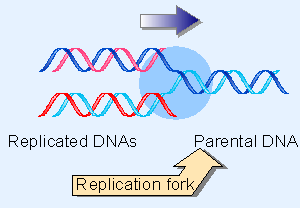 |
Figure 1.11 The replication fork is the region of DNA in which there is a transition from the unwound parental duplex to the newly replicated daughter duplexes. |
The helical structure of a molecule of DNA engaged in replication is illustrated in Figure 1.11. The nonreplicated region consists of the parental duplex, opening into the replicated region where the two daughter duplexes have formed. The double helical structure is disrupted at the junction between the two regions, called the replication fork. Replication involves movement of the replication fork along the parental DNA, so there is a continuous unwinding of the parental strands and rewinding into daughter duplexes.
The synthesis of nucleic acids is catalyzed by specific enzymes, which recognize the template and undertake the task of catalyzing the addition of subunits to the polynucleotide chain that is being synthesized. The enzymes are named according to the type of chain that is synthesized: DNA polymerases synthesize DNA, and RNA polymerases synthesize RNA.
Degradation of nucleic acids also requires specific enzymes: deoxyribonucleases (DNAases) degrade DNA, and ribonucleases (RNAases) degrade RNA. The nucleases fall into the general classes of exonucleases and endonucleases. Endonucleases cut individual bonds within RNA or DNA molecules, generating discrete fragments. They are involved in cutting reactions. Exonucleases remove residues one at a time from the end of the molecule, generating mononucleotides. They are involved in trimming reactions.
| Research | |
| Meselson, M. and Stahl, F. W. (1958). The replication of DNA in E. coli. Proc. Nat. Acad. Sci. USA 44, 671-682. | |