4. Ribozymes have various catalytic activities
23.3 Group I introns form a characteristic secondary structure |
 |
Figure 23.4 Group I introns have a common secondary structure that is formed by 9 base paired regions. The sequences of regions P4 and P7 are conserved, and identify the individual sequence elements P, Q, R, and S. P1 is created by pairing between the end of the left exon and the IGS of the intron; a region between P7 and P9 pairs with the 3' end of the intron. |
All group I introns can be organized into a characteristic secondary structure, with 9 helices (P1-P9). Figure 23.4 shows a model for the secondary structure of the Tetrahymena intron. Two of the base-paired regions are generated by pairing between conserved sequence elements that are common to group I introns. P4 is constructed from the sequences P and Q;, P7 is formed from sequences R and S. The other base-paired regions vary in sequence in individual introns. Mutational analysis identifies an intron "core," containing P3, P4, P6, and P7, which provides the minimal region that can undertake a catalytic reaction. The lengths of group I introns vary widely, and the consensus sequences are located a considerable distance from the actual splice junctions
Some of the pairing reactions are directly involved in bringing the splice junctions into a conformation that supports the enzymatic reaction. P1 includes the 3′ end of the left exon. The sequence within the intron that pairs with the exon is called the IGS, or internal guide sequence. (Its name reflects the fact that originally the region immediately 3′ to the IGS sequence shown in the figure was thought to pair with the 3′ splice junction, thus bringing the two junctions together. This interaction may occur, but does not seem to be essential.) A very short sequence, sometimes as short as 2 bases, between P7 and P9, base pairs with the sequence that immediately precedes the reactive G (position 414 in Tetrahymena) at the 3′ end of the intron (Michel and Wetshof, 1990).
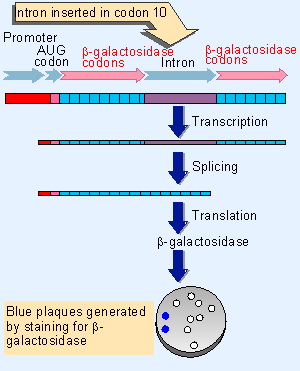 |
Figure 23.5 Placing the Tetrahymena intron within the b-galactosidase coding sequence creates an assay for self-splicing in E. coli. Synthesis of b-galactosidase can be tested by adding a compound that is turned blue by the enzyme. The sequence is carried by a bacteriophage, so the presence of blue plaques indicates successful splicing. |
The importance of base pairing in creating the necessary core structure in the RNA is emphasized by the properties of cis-acting mutations that prevent splicing of group I introns. Such mutations have been isolated for the mitochondrial introns through mutants that cannot remove an intron in vivo, and they have been isolated for the Tetrahymena intron by transferring the splicing reaction into a bacterial environment. The construct shown in Figure 23.5 allows the splicing reaction to be followed in E. coli. The self-splicing intron is placed at a location that interrupts the tenth codon of the β-galactosidase coding sequence. The protein can therefore be successfully translated from an RNA only after the intron has been removed.
The synthesis of β-galactosidase in this system indicates that splicing can occur in conditions quite distant from those prevailing in Tetrahymena or even in vitro. One interpretation of this result is that self-splicing can occur in the bacterial cell. Another possibility is that there are bacterial proteins that assist the reaction.
Using this assay, we can introduce mutations into the intron to see whether they prevent the reaction. Mutations in the group I consensus sequences that disrupt their base pairing stop splicing. The mutations can be reverted by making compensating changes that restore base pairing.
Mutations in the corresponding consensus sequences in mitochondrial group I introns have similar effects. A mutation in one consensus sequence may be reverted by a mutation in the complementary consensus sequence to restore pairing; for examples, mutations in the R consensus can be compensated by mutations in the S consensus.
Together these results suggest that the group I splicing reaction depends on the formation of secondary structure between pairs of consensus sequences within the intron. The principle established by this work is that sequences distant from the splice junctions themselves are required to form the active site that makes self-splicing possible (Burke et al., 1986).
| Research | |
| Burke, J. M. et al. (1986). Role of conserved sequence elements 9L and 2 in self-splicing of the Tetrahymena ribosomal RNA precursor. Cell 45, 167-176. | |
| Michel, F. and Wetshof, E. (1990). Modeling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216, 585-610. | |