3. DNA is coiled in arrays of nucleosomes
19.3 DNA is coiled in arrays of nucleosomes |
| Key terms defined in this section |
| Core DNA is the 146 bp of DNA contained on a core particle. Core particle is a digestion product of the nucleosome that retains the histone octamer and has 146 bp of DNA; its structure appears similar to that of the nucleosome itself. Linker DNA is all DNA contained on a nucleosome in excess of the 146 bp core DNA. |
 |
Figure 19.7 Micrococcal nuclease digests chromatin in nuclei into a multimeric series of DNA bands that can be separated by gel electrophoresis. Photograph kindly provided by Markus Noll. |
When chromatin is digested with the enzyme micrococcal nuclease, the DNA is cleaved into integral multiples of a unit length. Fractionation by gel electrophoresis reveals the "ladder" presented in Figure 19.7. Such ladders extend for ~10 steps, and the unit length, determined by the increments between successive steps, is ~200 bp.
 |
Figure 19.8 Each multimer of nucleosomes contains the appropriate number of unit lengths of DNA. Photograph kindly provided by John Finch. |
Figure 19.8 shows that the ladder is generated by groups of nucleosomes. When nucleosomes are fractionated on a sucrose gradient, they give a series of discrete peaks that correspond to monomers, dimers, trimers, etc. When the DNA is extracted from the individual fractions and electrophoresed, each fraction yields a band of DNA whose size corresponds with a step on the micrococcal nuclease ladder. The monomeric nucleosome contains DNA of the unit length, the nucleosome dimer contains DNA of twice the unit length, and so on (Finch et al., 1977).
So each step on the ladder represents the DNA derived from a discrete number of nucleosomes. We therefore take the existence of the 200 bp ladder in any chromatin to indicate that the DNA is organized into nucleosomes. The micrococcal ladder is generated when only ~2% of the DNA in the nucleus is rendered acid-soluble (degraded to small fragments) by the enzyme. So a small proportion of the DNA is specifically attacked; it must represent especially susceptible regions.
When chromatin is spilled out of nuclei, we often see a series of nucleosomes connected by a thread of free DNA (the beads on a string). However, the need for tight packaging of DNA in vivo suggests that probably there is usually little (if any) free DNA.
This view is confirmed by the fact that >90% of the DNA of chromatin can be recovered in the form of the 200 bp ladder. Almost all DNA must therefore be organized in nucleosomes. In their natural state, nucleosomes are likely to be closely packed, with DNA passing directly from one to the next. Free DNA is probably generated by the loss of some histone octamers during isolation.
The length of DNA present in the nucleosome varies somewhat from the "typical" value of 200 bp. The chromatin of any particular cell type has a characteristic average value ( 5 bp). The average most often is between 180 and 200, but there are extremes as low as 154 bp (in a fungus) or as high as 260 bp (in a sea urchin sperm). The average value may be different in individual tissues of the adult organism. And there can be differences between different parts of the genome in a single cell type. Variations from the genome average include tandemly repeated sequences, such as clusters of 5S RNA genes.
A common structure underlies the varying amount of DNA that is contained in nucleosomes of different sources. The association of DNA with the histone octamer forms a core particle containing 146 bp of DNA, irrespective of the total length of DNA in the nucleosome. The variation in total length of DNA per nucleosome is superimposed on this basic core structure.
The core particle is defined by the effects of micrococcal nuclease on the nucleosome monomer. The initial reaction of the enzyme is to cut between nucleosomes, but if it is allowed to continue after monomers have been generated, then it proceeds to digest some of the DNA of the individual nucleosome. This occurs by a reaction in which DNA is "trimmed" from the ends of the nucleosome.
 |
Figure 19.9 Micrococcal nuclease reduces the length of nucleosome monomers in discrete steps. Photograph kindly provided by Roger Kornberg. |
The length of the DNA is reduced in discrete steps, as shown in Figure 19.9. With rat liver nuclei, the nucleosome monomers initially have 205 bp of DNA. Then some monomers are found in which the length of DNA has been reduced to ~165 bp. Finally this is reduced to the length of the DNA of the core particle, 146 bp. (The core is reasonably stable, but continued digestion generates a limit digest, in which the longest fragments are the 146 bp DNA of the core, while the shortest are as small as 20 bp.)
This analysis suggests that the nucleosomal DNA can be divided into two regions:
- Core DNA has an invariant length of 146 bp, and is relatively resistant to digestion by nucleases.
- Linker DNA comprises the rest of the repeating unit. Its length varies from as little as 8 bp to as much as 114 bp per nucleosome.
 |
Figure 19.10 Microccocal nuclease initially cleaves between nucleosomes. Mononucleosomes typically have ~200 bp DNA. End-trimming reduces the length of DNA first to ~165 bp, and then generates core particles with 146 bp. |
The sharp size of the band of DNA generated by the initial cleavage with micrococcal nuclease suggests that the region immediately available to the enzyme is restricted. It represents only part of each linker. (If the entire linker DNA were susceptible, the band would range from 146 bp to >200 bp.) But once a cut has been made in the linker DNA, the rest of this region becomes susceptible, and it can be removed relatively rapidly by further enzyme action. The connection between nucleosomes is represented in Figure 19.10.
Core particles have properties similar to those of the nucleosomes themselves, although they are smaller. Their shape and size are similar to nucleosomes, which suggests that the essential geometry of the particle is established by the interactions between DNA and the protein octamer in the core particle. Because core particles are more readily obtained as a homogeneous population, they are often used for structural studies in preference to nucleosome preparations. (Nucleosomes tend to vary because it is difficult to obtain a preparation in which there has been no end-trimming of the DNA.)
What is the physical nature of the core and the linker regions? These terms are operational definitions that describe the regions in terms of their relative susceptibility to nuclease treatment. This description does not make any implication about their actual structure. In fact, the path of DNA on the histone octamer appears to be continuous. It takes 165 bp to make the two turns around the octamer. This is an invariant feature of nucleosomes. The transition from one nucleosome to the next is made within the additional length of DNA, and there could be differences in the path in this region depending on the length of DNA per nucleosome.
The existence of linker DNA depends on factors extraneous to the four core histones. Reconstitution experiments in vitro show that histones have an intrinsic ability to organize DNA into core particles, but do not form nucleosomes with the proper unit length. The degree of supercoiling of the DNA is an important factor. Histone H1 and/or nonhistone proteins influence the length of linker DNA associated with the histone octamer in a natural series of nucleosomes. And "assembly proteins" that are not part of the nucleosome structure are involved in vivo in constructing nucleosomes from histones and DNA (see later).
Where is histone H1 located? The H1 is lost during the degradation of nucleosome monomers. It can be retained on monomers that still have 165 bp of DNA; but is always lost with the final reduction to the 146 bp core particle. This suggests that H1 could be located in the region of the linker DNA immediately adjacent to the core DNA.
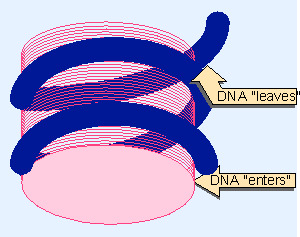 |
Figure 19.4 The nucleosome may be a cylinder with DNA organized into two turns around the surface. |
If H1 is located at the linker, it could "seal" the DNA in the nucleosome by binding at the point where the nucleic acid enters and leaves (see Figure 19.4). The idea that H1 lies in the region joining adjacent nucleosomes is consistent with old results that H1 is removed the most readily from chromatin, and that H1-depleted chromatin is more readily "solubilized". And it is easier to obtain a stretched-out fiber of beads on a string when the H1 has been removed (Shen et al., 1995).
| Research | |
| Finch, J. T. et al. (1977). Structure of nucleosome core particles of chromatin. Nature 269, 29-36. | |
| Shen, X. et al. (1995). Linker histones are not esssential and affect chromatin condensation in vivo. Cell 82, 47-56. | |