8. The eukaryotic chromosome as a segregation device
18.8 The eukaryotic chromosome as a segregation device |
| Key terms defined in this section |
| Acentric fragment of a chromosome (generated by breakage) lacks a centromere and is lost at cell division. Centromere is a constricted region of a chromosome that includes the site of attachment (the kinetochore) to the mitotic or meiotic spindle. Kinetochore is the structural feature of the chromosome to which microtubules of the mitotic spindle attach. Its location determines the centromneric region. MTOC (microtubule organizing center) is a region from which microtubules emanate. The major MTOCs in a mitotic cell are the centrosomes. |
During mitosis, the sister chromatids move to opposite poles of the cell. Their movement depends on the attachment of the chromosome to microtubules, which are connected at their other end to the poles. (The microtubules comprise a cellular filamentous system, reorganized at mitosis so that they connect the chromosomes to the poles of the cell.) The sites in the two regions where microtubule ends are organized Xin the vicinity of the centrioles at the poles and at the chromosomes Xare called MTOCs (microtubule organizing centers).
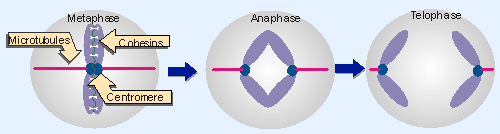 |
Figure 18.19 Chromosomes are pulled to the poles via microtubules that attach to their kinetochores. They sister chromatids are held together until anaphase by glue proteins (cohesins). The centromere is shown here in the middle of the chromosome, but can be located anywhere along its length, including close to the end (acrocentric) and at the end (telocentric). Animated figure |
Figure 18.19 illustrates the separation of sister chromatids as mitosis proceeds from metaphase to telophase. The region of the chromosome that is responsible for its segregation at mitosis and meiosis is called the centromere. The centromeric region on each sister chromatid is pulled by microtubules to the opposite pole. Opposing this motive force, "glue" proteins called cohesins hold the sister chromatids together . Initially the sister chromatids separate at their centomeres, and then thet are released completely from one another during anaphase when the cohesins are degraded (the cohesins are discussed in more detail in 27.9 Protein degradation is important in mitosis).
 |
Figure 18.9 The sister chromatids of a mitotic pair each consist of a fiber (~30 nm in diameter) compactly folded into the chromosome. Photograph kindly provided by E. J. DuPraw. |
The term "centromere" historically has been used in both the functional and structural sense to describe the feature of the chromosome responsible for its movement. The centromere is pulled toward the pole during mitosis, and the attached chromosome is dragged along behind, as it were. The chromosome therefore provides a device for attaching a large number of genes to the apparatus for division. It contains the site at which the sister chromatids are held together prior to the separation of the individual chromosomes. This shows as a constricted region connecting all four chromosome arms, as in the photograph of Figure 18.9, which shows the sister chromatids at the metaphase stage of mitosis.
The centromere is essential for segregation, as shown by the behavior of chromosomes that have been broken. A single break generates one piece that retains the centromere, and another, an acentric fragment, that lacks it. The acentric fragment does not become attached to the mitotic spindle; and as a result it fails to be included in either of the daughter nuclei.
(When chromosome movement relies on discrete centromeres, there can be only one centromere per chromosome. When translocations generate chromosomes with more than one centromere, aberrant structures form at mitosis, since the two centromeres on the same sister chromatid can be pulled toward different poles, breaking the chromosome. However, in some species the centromeres are "diffuse," which creates a different situation. Only discrete centromeres have been analyzed at the molecular level.)
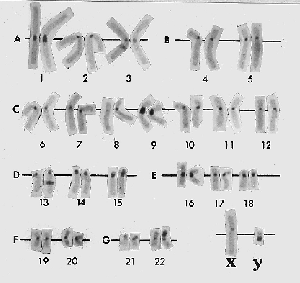 |
Figure 18.20 C-banding generates intense staining at the centromeres of all chromosomes. Photograph kindly provided by Lisa Shaffer. |
The regions flanking the centromere often are rich in satellite DNA sequences and display a considerable amount of heterochromatin. Because the entire chromosome is condensed, centromeric heterochromatin is not immediately evident in mitotic chromosomes. However, it can be visualized by a technique called C-banding. In the example of Figure 18.20, all the centromeres show as darkly staining regions. Although it is common, heterochromatin cannot be identified around every known centromere, which suggests that it is unlikely to be essential for the division mechanism.
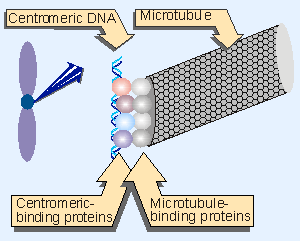 |
Figure 18.21 The centromere is identified by a DNA sequence that binds specific proteins. These proteins do not themselves bind to microtubules, but establish the site a which the microtubule-binding proteins in turn bind. |
The region of the chromosome at which the centromere forms is defined by DNA sequences (although the sequences have been defined in only a very small number of cases). The centromeric DNA binds specific proteins that are responsible for establishing the structure that attaches the chromosome to the microtubules. This structure is called the kinetochore. It is a darkly staining fibrous object of diameter or length ~400 nm. The kinetochore provides the MTOC on a chromosome (for review see Hyman and Sorger, 1995). Figure 18.21 shows the hierarchy of organization that connects centromericDNA to the microtubule. Proteins bound to the centromeric DNA bind other proteins that bind to microtubule. It remains to be seen exactly which proteins are involved and how they are related to the visible structure of the kinetochore.
If a centromeric sequence of DNA is responsible for segregation, any molecule of DNA possessing this sequence should move properly at cell division, while any DNA lacking it will fail to segregate. This prediction has been used to isolate centromeric DNA in the yeast, S. cerevisiae. Yeast chromosomes do not display visible kinetochores comparable to those of higher eukaryotes, but otherwise divide at mitosis and segregate at meiosis by the same mechanisms.
Genetic engineering has produced plasmids of yeast that are replicated like chromosomal sequences (see 12 The replicon). However, they are unstable at mitosis and meiosis, disappearing from a majority of the cells because they segregate erratically. Fragments of chromosomal DNA have been isolated by virtue of their ability to confer mitotic stability on these plasmids.
A CEN fragment is defined by its ability to confer stability upon such a plasmid. By reducing the sizes of the fragments that are incorporated into the plasmid, the minimum region necessary for mitotic centromeric function can be identified. Deletions and other changes can be made to investigate the features involved in centromeric function.
Another way to use the availability of the centromeric sequences is to modify them in vitro and then reintroduce them into the yeast cell, where they replace the corresponding centromere on the chromosome. This allows the sequences required for CEN function to be defined directly in the context of the chromosome (for review see Blackburn and Szostak, 1984).
A CEN fragment derived from one chromosome can replace the centromere of another chromosome with no apparent consequence. This result suggests that centromeres are interchangeable. They are used simply to attach the chromosome to the spindle, and play no role in distinguishing one chromosome from another.
The sequences required for centromeric function fall within a stretch of ~120 bp. The centromeric region is packaged into a nuclease-resistant structure, and it binds a single microtubule. We may therefore look to the S. cerevisiae centromeric region to identify proteins that bind centromeric DNA and proteins that connect the chromosome to the spindle.
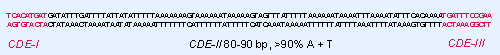 |
Figure 18.22 Three conserved regions can be identified by the sequence homologies between yeast CEN elements. |
Three types of sequence element may be distinguished in the CEN region, as summarized in Figure 18.22:
- CDE-I is a sequence of 9 bp that is conserved with minor variations at the left boundary of all centromeres.
- CDE-II is a >90% A PT-rich sequence of 80 V90 bp found in all centromeres; its function could depend on its length rather than exact sequence. Its constitution is reminiscent of some short tandemly repeated (satellite) DNAs in higher eukaryotes (see 4 Clusters and repeats). Its base composition may cause some characteristic distortions of the DNA double helical structure.
- CDE-III is an 11 bp sequence highly conserved at the right boundary of all centromeres. Sequences on either side of the element are less well conserved, and may also be needed for centromeric function. (CDE-III could be longer than 11 bp if it turns out that the flanking sequences are essential.)
Mutations in CDE-I or CDE-II reduce but do not inactivate centromere function, but point mutations in the central CCG of CDE-III completely inactivate the centromere (for review see Clarke and Carbon, 1985).
Can we identify proteins that are necessary for the function of CEN sequences? CDE-I is bound by the homodimer CBF1; this interaction is not essential for centromere function, but in its absence the fidelity of segregation chromosome is reduced ~10 . A 240 kD complex of several proteins, called CBF3, binds to CDE-III. This interaction is essential for centromeric function. Other proteins may bind to the CBF1 and CBF3 complexes to assemble the full centromeric structure. The complex is integrated into the chromosomal structure (see 19 Nucleosomes).
Mutations in the components of the genes coding for CBF-III block chromosome movement at mitosis; and the protein complex has a microtubule-based motor activity Xit is able to move itself, and objects attached to it, such as chromosomes, along microtubules. Comparable proteins with sequences related to known motor activities have been found at eukaryotic centromeres. Taken together, these observations suggest that a protein complex with motor activity may connect the centromeric region of a chromosome to microtubules, and contribute to movement on the mitotic spindle. The discovery of the CBF-III complex may be the prelude to characterizing the connection between the centromere and the apparatus for chromosome segregation (for review see Schulman and Bloom, 1991).
Attempts to characterize functional centromeres from the yeast S. pombe have been less successful. They cannot be isolated by ability to confer stability on plasmids. However, S. pombe has only 3 chromosomes, and the region containing each centromere has been identified by deleting most of the sequences of each chromosome to create a stable minichromosome. This approach locates the centromeres within regions of 40 V100 kb that consist largely or entirely of repetitious DNA. It is not clear how much of each of these rather long regions is required for chromosome segregation at mitosis and meiosis.
The significance of the difference between the short centromeric regions in S. cerevisiae and the long regions in S. pombe is not clear. The common feature is that the DNA consists of noncoding sequences that are repetitive.
Attempts to localize centromeric functions in Drosophila chromosomes suggest that they are dispersed in a large region, consisting of 200 V600 kb. The large size of this type of centromere suggests that it is likely to contain several separate specialized functions, including sequences required for kinetochore assembly, sister chromatid pairing, etc.
The size of the centromere in Arabidospsis is comparable (Copenhaver et al., 1999). Each of the 5 chromosomes has a centromeric region in which recombination is very largely suppressed. This region occupies >500 kb. Clearly it includes the centromere, but we have no direct information as to how much of it is required. There are expressed genes within these regions, which casts some doubt on whether the entire region is part of the centromere. At the center of the region is a series of 180 bp repeats; this is the type of structure generally associated with centromeres. It is too early to say how these structures relate to centromeric function.
The primary motif comprising the heterochromatin of primate centromeres is the α satellite DNA, which consists of tandem arrays of a 170 bp repeating unit. There is significant variation between individual repeats, although those at any centromere tend to be better related to one another than to members of the family in other locations. It is clear that the sequences required for centromeric function reside within the blocks of α satellite DNA, but it is not clear whether the α satellite sequences themselves provide this function, or whether other sequences are embedded within the α satellite arrays.
This section updated 1-10-2000
| Reviews | |
| Blackburn, E. H. and Szostak, J. W. (1984). The molecular structure of centromeres and telomeres. Ann. Rev. Biochem 53, 163-194. | |
| Clarke, L. and Carbon, J. (1985). The structure and function of yeast centromeres. Ann. Rev. Genet. 19, 29-56. | |
| Hyman, A. A. and Sorger, P. K. (1995). Structure and function of kinetochores in budding yeast. Ann. Rev. Cell Dev. Biol. 11, 471-495. | |
| Schulman, I. and Bloom, K. S. (1991). Centromeres: an integrated protein/DNA complex required for chromosome movement. Ann. Rev. Cell Biol. 7, 311-336. | |
| Research | |
| Copenhaver, G. P. et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres.. Science 286, 2468-2474. | |