4. DNA polymerases control the fidelity of replication
13.4 DNA polymerases control the fidelity of replication |
| Key terms defined in this section |
| Proofreading refers to any mechanism for correcting errors in protein or nucleic acid synthesis that involves scrutiny of individual units after they have been added to the chain. |
The fidelity of replication poses the same sort of problem we have encountered already in considering (for example) the accuracy of translation. It relies on the specificity of base pairing. Yet when we consider the interactions involved in base pairing, we would expect errors to occur with a frequency of ~10 V3 per base pair replicated. The actual rate in bacteria seems to be ~10 V8 V10 V10. This corresponds to ~1 error per genome per 1000 bacterial replication cycles, or ~10 V6 per gene per generation.
We can divide the errors that DNA polymerase makes during replication into two classes. Substitutions occur when the wrong (improperly paired) nucleotide is incorporated. The error level is determined by the efficiency of proofreading. Frameshifts occur when an extra nucleotide is inserted or omitted. Fidelity with regard to frameshifts is affected by the processivity of the enzyme: the tendency to remain on a single template rather than to dissociate and reassociate. This is particularly important for the replication of a homopolymeric stretch, for example, a long sequence of dTn:dAn, in which "replication slippage" can change the length of the homopolymeric run. As a general rule, increased processivity reduces the likelihood of such events. In multimeric DNA polymerases, processivity is usually increased by a particular subunit that is not needed for catalytic activity per se.
DNA polymerase might improve the specificity of complementary base selection at either (or both) of two stages:
- It could scrutinize the incoming base for the proper complementarity with the template base; for example, by specifically recognizing matching chemical features. This would be a presynthetic error control.
- Or it could scrutinize the base pair after the new base has been added to the chain, and, in those cases in which a mistake has been made, remove the most recently added base. This would be a proofreading control.
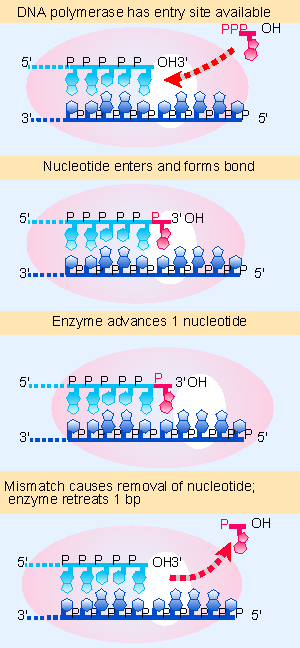 |
Figure 13.6 Bacterial DNA polymerases scrutinize the base pair at the end of the growing chain and excise the nucleotide added in the case of a misfit. |
All of the bacterial enzymes possess a 3′ V5′ exonucleolytic activity that proceeds in the reverse direction from DNA synthesis. This provides the proofreading function illustrated diagrammatically in Figure 13.6. In the chain elongation step, a precursor nucleotide enters the position at the end of the growing chain. A bond is formed. The enzyme moves one base pair farther, ready for the next precursor nucleotide to enter. If a mistake has been made, the enzyme moves backward, excising the last base that was added, and creating a site for a replacement precursor nucleotide to enter.
Different DNA polymerases handle the relationship between the polymerizing and proofreading activities in different ways. In some cases, the activities are part of the same protein subunit, but in others they are contained in different subunits. Each DNA polymerase has a characteristic error rate that is reduced by its proofreading activity. Proofreading typically decreases the error rate in replication from ~10 V5 to ~10 V7.