15. Plasmid incompatibility is connected with copy number
12.15 Plasmid incompatibility is connected with copy number |
| Key terms defined in this section |
| Compatibility group of plasmids contains members unable to coexist in the same bacterial cell. |
The phenomenon of plasmid incompatibility is related to the regulation of plasmid copy number and segregation. A compatibility group is defined as a set of plasmids whose members are unable to coexist in the same bacterial cell. The reason for their incompatibility is that they cannot be distinguished from one another at some stage that is essential for plasmid maintenance. DNA replication and segregation are stages at which this may apply (for review see Scott, 1984; Nordstrom and Austin, 1989).
The negative control model for plasmid incompatibility follows the idea that copy number control is achieved by synthesizing a repressor that measures the concentration of origins. (Formally this is the same as the titration model for regulating replication of the bacterial chromosome.)
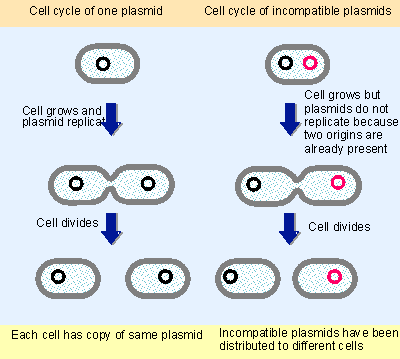 |
Figure 12.33 Two plasmids are incompatible (they belong to the same compatibility group) if their origins cannot be distinguished at the stage of initiation. The same model could apply to segregation. |
The introduction of a new origin in the form of a second plasmid of the same compatibility group mimics the result of replication of the resident plasmid; two origins now are present. So any further replication is prevented until after the two plasmids have been segregated to different cells to create the correct prereplication copy number as illustrated in Figure 12.33.
A similar effect would be produced if the system for segregating the products to daughter cells could not distinguish between two plasmids. For example, if two plasmids have the same cis-acting partition sites, competition between them would ensure that they would be segregated to different cells, and therefore could not survive in the same line.
Viewed in teleological terms, plasmids are selfish. Having obtained possession of a bacterium, the resident plasmid will seek to prevent any other plasmid of the same type from establishing residence. Plasmid incompatibility is a major device used to establish these territorial rights.
The presence of a member of one compatibility group does not directly affect the survival of a plasmid belonging to a different group. Only one replicon of a given compatibility group (of a single Vcopy plasmid) can be maintained in the bacterium, but it does not interact with replicons of other compatibility groups (although in limiting conditions they compete for lebensraum).
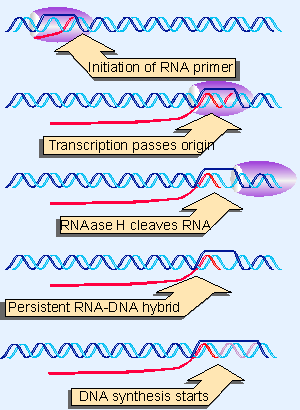 |
Figure 12.34 Replication of ColE1 DNA is initiated by cleaving the primer RNA to generate a 3 F-OH end. The primer forms a persistent hybrid in the origin region. |
The best characterized copy number and incompatibility system is that of the plasmid ColE1, a multicopy plasmid that is maintained at a steady level of ~20 copies per E. coli cell. The system for maintaining the copy number depends on the mechanism for initiating replication at the ColE1 origin, as illustrated in Figure 12.34.
Replication starts with the transcription of an RNA that initiates 555 bp upstream of the origin. Transcription continues through the origin. The enzyme RNAase H (whose name reflects its specificity for a substrate of RNA hybridized with DNA) cleaves the transcript at the origin. This generates a 3′ VOH end that is used as the "primer" at which DNA synthesis is initiated (the use of primers is discussed in more detail in 13 DNA replication). The primer RNA forms a persistent hybrid with the DNA. Pairing between the RNA and DNA occurs just upstream of the origin (around position V20) and also farther upstream (around position V265).
Two regulatory systems exert their effects on the RNA primer. One involves synthesis of an RNA complementary to the primer; the other involves a protein coded by a nearby locus.
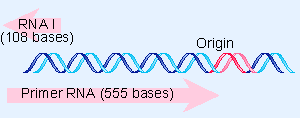 |
Figure 12.35 The sequence of RNA I is complementary to the 5 F region of primer RNA. |
The regulatory species RNA I is a molecule of ~108 bases, coded by the opposite strand from that specifying primer RNA. The relationship between the primer RNA and RNA I is illustrated in Figure 12.35. The RNA I molecule is initiated within the primer region and terminates close to the site where the primer RNA initiates. So RNA I is complementary to the 5′ Vterminal region of the primer RNA. Base pairing between the two RNAs controls the availability of the primer RNA to initiate a cycle of replication (Tomizawa and Itoh, 1981; Masukata and Tomizawa, 1990).
An RNA molecule such as RNA I that functions by virtue of its complementarity with another RNA coded in the same region is called a countertranscript. This type of mechanism, of course, is another example of the use of antisense RNA (see 10 The operon).
Mutations that reduce or eliminate incompatibility between plasmids can be obtained by selecting plasmids of the same group for their ability to coexist. Incompatibility mutations in ColE1 map in the region of overlap between RNA I and primer RNA. Because this region is represented in two different RNAs, either or both might be involved in the effect.
When RNA I is added to a system for replicating ColE1 DNA in vitro, it inhibits the formation of active primer RNA. But the presence of RNA I does not inhibit the initiation or elongation of primer RNA synthesis. This suggests that RNA I prevents RNAase H from generating the 3′ end of the primer RNA. The basis for this effect lies in base pairing between RNA I and primer RNA.
Both RNA molecules have the same potential secondary structure in this region, with three duplex hairpins terminating in single Vstranded loops. Mutations reducing incompatibility are located in these loops, which suggests that the initial step in base pairing between RNA I and primer RNA is contact between the unpaired loops.
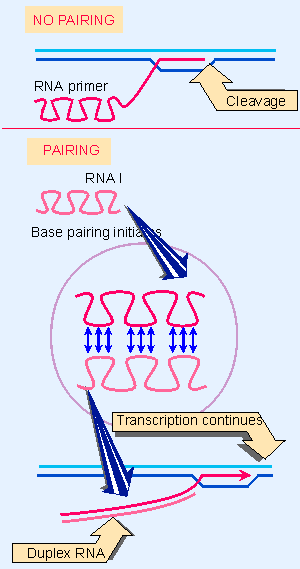 |
Figure 12.36 Base pairing with RNA I may change the secondary structure of the primer RNA sequence and thus prevent cleavage from generating a 3 F-OH end. |
How does pairing with RNA I prevent cleavage to form primer RNA? A model is illustrated in Figure 12.36. In the absence of RNA I, the primer RNA forms its own secondary structure (involving loops and stems). But when RNA I is present, the two molecules pair, and become completely double Vstranded for the entire length of RNA I. The new secondary structure prevents the formation of the primer, probably by affecting the ability of the RNA to form the persistent hybrid.
The model resembles the mechanism involved in attenuation of transcription, in which the alternative pairings of an RNA sequence permit or prevent formation of the secondary structure needed for termination by RNA polymerase (see 10 The operon). The action of RNA I is exercised by its ability to affect distant regions of the primer precursor.
Formally, the model is equivalent to postulating a control circuit involving two RNA species. A large RNA primer precursor is a positive regulator, needed to initiate replication. The small RNA I is a negative regulator, able to inhibit the action of the positive regulator.
In its ability to act on any plasmid present in the cell, RNA I provides a repressor that prevents newly introduced DNA from functioning, analogous to the role of the lambda lysogenic repressor (see 11 Phage strategies). Instead of a repressor protein that binds the new DNA, an RNA binds the newly synthesized precursor to the RNA primer.
Binding between RNA I and primer RNA can be influenced by the Rom protein, coded by a gene located downstream of the origin. The Rom protein enhances binding between RNA I and primer RNA transcripts of >200 bases. The result is to inhibit formation of the primer.
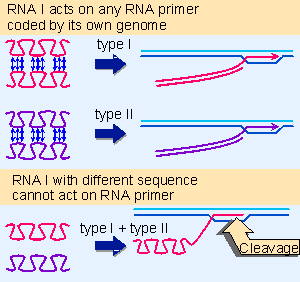 |
Figure 12.37 Mutations in the region coding for RNA I and the primer precursor need not affect their ability to pair; but they may prevent pairing with the complementary RNA coded by a different plasmid. |
How do mutations in the RNAs affect incompatibility? Figure 12.37 shows the situation when a cell contains two types of RNA I/primer RNA sequence. The RNA I and primer RNA made from each type of genome can interact, but RNA I from one genome does not interact with primer RNA from the other genome. This situation would arise when a mutation in the region that is common to RNA I and primer RNA occurred at a location that is involved in the base pairing between them. Each RNA I would continue to pair with the primer RNA coded by the same plasmid, but might be unable to pair with the primer RNA coded by the other plasmid. This would cause the original and the mutant plasmids to behave as members of different compatibility groups.
| Reviews | |
| Nordstrom, K. and Austin, S. J. (1989). Mechanisms that contribute to the stable segregation of plasmids. Ann. Rev. Genet. 23, 37-69. | |
| Scott, J. R. (1984). Regulation of plasmid replication. Microbiol. Rev. 48, 1-23. | |
| Research | |
| Masukata, H. and Tomizawa, J. (1990). A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 62, 331-338. | |
| Tomizawa, J.-I. and Itoh, T. (1981). Plasmid ColE1 incompatibility determined by interaction of RNA with primer transcript. Proc. Nat. Acad. Sci. USA 78, 6096-6100. | |